Nuclear recycling of the pre-60S ribosomal subunit-associated factor Arx1 depends on Rei1 in Saccharomyces cerevisiae
- PMID: 16648468
- PMCID: PMC1489010
- DOI: 10.1128/MCB.26.10.3718-3727.2006
Nuclear recycling of the pre-60S ribosomal subunit-associated factor Arx1 depends on Rei1 in Saccharomyces cerevisiae
Abstract
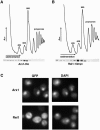
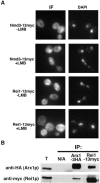
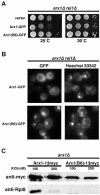
Arx1 and Rei1 are found on late pre-60S ribosomal particles containing the export adaptor Nmd3. Arx1 is related to methionine aminopeptidases (MetAPs), and Rei1 is a C2H2 zinc finger protein whose function in ribosome biogenesis has not been previously characterized. Arx1 and Rei1 localized predominately to the nucleus and cytoplasm, respectively, but could be coimmunoprecipitated, suggesting that they are transiently in the same 60S complex. arx1delta mutants showed a modest accumulation of 60S subunits in the nucleus, suggesting that Arx1 enhances 60S export. Deletion of REI1 led to cold sensitivity and redistribution of Arx1 to the cytoplasm, where it remained bound to free 60S subunits. However, deletion of ARX1 or the fusion of enhanced GFP (eGFP) to Rpl25 suppressed the cold sensitivity of an rei1delta mutant. The presence of eGFP on Rpl25 or its neighboring protein Rpl35 reduced the binding of Arx1 to 60S subunits, suggesting that Arx1 binds to 60S subunits in the vicinity of the exit tunnel. Mutations in Arx1 that disrupted its binding to 60S also suppressed an rei1delta mutant and restored the normal nuclear localization of Arx1. These results indicate that the cold sensitivity of rei1delta cells is due to the persistence of Arx1 on 60S subunits in the cytoplasm. Furthermore, these results suggest that Rei1 is needed for release of Arx1 from nascent 60S subunits after export to the cytoplasm but not for the subsequent nuclear import of Arx1.
Figures

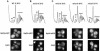



Similar articles
-
Arx1 is a nuclear export receptor for the 60S ribosomal subunit in yeast.Mol Biol Cell. 2008 Feb;19(2):735-44. doi: 10.1091/mbc.e07-09-0968. Epub 2007 Dec 12. Mol Biol Cell. 2008. PMID: 18077551 Free PMC article.
-
Functional redundancy of yeast proteins Reh1 and Rei1 in cytoplasmic 60S subunit maturation.Mol Cell Biol. 2009 Jul;29(14):4014-23. doi: 10.1128/MCB.01582-08. Epub 2009 May 11. Mol Cell Biol. 2009. PMID: 19433447 Free PMC article.
-
Cryo-EM structures of Arx1 and maturation factors Rei1 and Jjj1 bound to the 60S ribosomal subunit.Nat Struct Mol Biol. 2012 Dec;19(12):1228-33. doi: 10.1038/nsmb.2425. Epub 2012 Nov 11. Nat Struct Mol Biol. 2012. PMID: 23142985
-
Nuclear export and cytoplasmic maturation of ribosomal subunits.FEBS Lett. 2007 Jun 19;581(15):2783-93. doi: 10.1016/j.febslet.2007.05.013. Epub 2007 May 11. FEBS Lett. 2007. PMID: 17509569 Review.
-
Nuclear export of ribosomal subunits.Trends Biochem Sci. 2002 Nov;27(11):580-5. doi: 10.1016/s0968-0004(02)02208-9. Trends Biochem Sci. 2002. PMID: 12417134 Review.
Cited by
-
N6-methyl-adenosine (m6A) in RNA: an old modification with a novel epigenetic function.Genomics Proteomics Bioinformatics. 2013 Feb;11(1):8-17. doi: 10.1016/j.gpb.2012.12.002. Epub 2012 Dec 21. Genomics Proteomics Bioinformatics. 2013. PMID: 23453015 Free PMC article. Review.
-
Ribosomal protein L35 is required for 27SB pre-rRNA processing in Saccharomyces cerevisiae.Nucleic Acids Res. 2010 Aug;38(15):5177-92. doi: 10.1093/nar/gkq260. Epub 2010 Apr 14. Nucleic Acids Res. 2010. PMID: 20392820 Free PMC article.
-
Plant Temperature Acclimation and Growth Rely on Cytosolic Ribosome Biogenesis Factor Homologs.Plant Physiol. 2018 Mar;176(3):2251-2276. doi: 10.1104/pp.17.01448. Epub 2018 Jan 30. Plant Physiol. 2018. PMID: 29382692 Free PMC article.
-
A molecular genetic toolbox for Yarrowia lipolytica.Biotechnol Biofuels. 2017 Jan 3;10:2. doi: 10.1186/s13068-016-0687-7. eCollection 2017. Biotechnol Biofuels. 2017. PMID: 28066508 Free PMC article.
-
Defining the pathway of cytoplasmic maturation of the 60S ribosomal subunit.Mol Cell. 2010 Jul 30;39(2):196-208. doi: 10.1016/j.molcel.2010.06.018. Mol Cell. 2010. PMID: 20670889 Free PMC article.
References
-
- Bassler, J., P. Grandi, O. Gadal, T. Lessmann, E. Petfalski, D. Tollervey, J. Lechner, and E. Hurt. 2001. Identification of a 60S preribosomal particle that is closely linked to nuclear export. Mol. Cell 8:517-529. - PubMed
-
- Beckmann, R., C. M. Spahn, N. Eswar, J. Helmers, P. A. Penczek, A. Sali, J. Frank, and G. Blobel. 2001. Architecture of the protein-conducting channel associated with the translating 80S ribosome. Cell 107:361-372. - PubMed
-
- Ceci, M., C. Gaviraghi, C. Gorrini, L. A. Sala, N. Offenhauser, P. C. Marchisio, and S. Biffo. 2003. Release of eIF6 (p27BBP) from the 60S subunit allows 80S ribosome assembly. Nature 426:579-584. - PubMed
-
- Ciufo, L. F., and J. D. Brown. 2000. Nuclear export of yeast signal recognition particle lacking Srp54p by the Xpo1p/Crm1p NES-dependent pathway. Curr. Biol. 10:1256-1264. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
