Furin-, ADAM 10-, and gamma-secretase-mediated cleavage of a receptor tyrosine phosphatase and regulation of beta-catenin's transcriptional activity
- PMID: 16648485
- PMCID: PMC1489012
- DOI: 10.1128/MCB.26.10.3917-3934.2006
Furin-, ADAM 10-, and gamma-secretase-mediated cleavage of a receptor tyrosine phosphatase and regulation of beta-catenin's transcriptional activity
Abstract
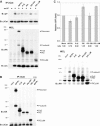
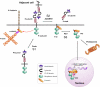
Several receptor protein tyrosine phosphatases (RPTPs) are cell adhesion molecules involved in homophilic interactions, suggesting that RPTP outside-in signaling is coupled to cell contact formation. However, little is known about the mechanisms by which cell density regulates RPTP function. We show that the MAM family prototype RPTPkappa is cleaved by three proteases: furin, ADAM 10, and gamma-secretase. Cell density promotes ADAM 10-mediated cleavage and shedding of RPTPkappa. This is followed by gamma-secretase-dependent intramembrane proteolysis of the remaining transmembrane part to release the phosphatase intracellular portion (PIC) from the membrane, thereby allowing its translocation to the nucleus. When cells were treated with leptomycin B, a nuclear export inhibitor, PIC accumulated in nuclear bodies. PIC is an active protein tyrosine phosphatase that binds to and dephosphorylates beta-catenin, an RPTPkappa substrate. The expression of RPTPkappa suppresses beta-catenin's transcriptional activity, whereas the expression of PIC increases it. Notably, this increase required the phosphatase activity of PIC. Thus, both isoforms have acquired opposing roles in the regulation of beta-catenin signaling. We also found that RPTPmu, another MAM family member, undergoes gamma-secretase-dependent processing. Our results identify intramembrane proteolysis as a regulatory switch in RPTPkappa signaling and implicate PIC in the activation of beta-catenin-mediated transcription.
Figures






References
-
- Bradykalnay, S. M., and N. K. Tonks. 1995. Protein tyrosine phosphatases as adhesion receptors. Curr. Opin. Cell Biol. 7:650-657. - PubMed
-
- Campan, M., M. Yoshizumi, N. G. Seidah, M. E. Lee, C. Bianchi, and E. Haber. 1996. Increased proteolytic processing of protein tyrosine phosphatase μ in confluent vascular endothelial cells—the role of pc5, a member of the subtilisin family. Biochemistry 35:3797-3802. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
