Evolution and expression of homeologous loci in Tragopogon miscellus (Asteraceae), a recent and reciprocally formed allopolyploid
- PMID: 16648586
- PMCID: PMC1526671
- DOI: 10.1534/genetics.106.057646
Evolution and expression of homeologous loci in Tragopogon miscellus (Asteraceae), a recent and reciprocally formed allopolyploid
Abstract
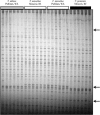
On both recent and ancient time scales, polyploidy (genome doubling) has been a significant evolutionary force in plants. Here, we examined multiple individuals from reciprocally formed populations of Tragopogon miscellus, an allotetraploid that formed repeatedly within the last 80 years from the diploids T. dubius and T. pratensis. Using cDNA-AFLPs followed by genomic and cDNA cleaved amplified polymorphic sequence (CAPS) analyses, we found differences in the evolution and expression of homeologous loci in T. miscellus. Fragment variation within T. miscellus, possibly attributable to reciprocal formation, comprised 0.6% of the cDNA-AFLP bands. Genomic and cDNA CAPS analyses of 10 candidate genes revealed that only one "transcript-derived fragment" (TDF44) showed differential expression of parental homeologs in T. miscellus; the T. pratensis homeolog was preferentially expressed by most polyploids in both populations. Most of the cDNA-AFLP polymorphisms apparently resulted from loss of parental fragments in the polyploids. Importantly, changes at the genomic level have occurred stochastically among individuals within the independently formed populations. Synthetic F(1) hybrids between putative diploid progenitors are additive of their parental genomes, suggesting that polyploidization rather than hybridization induces genomic changes in Tragopogon.
Figures


References
-
- Bachem, C. W. B., R. S. van der Hoeven, S. M. de Bruijn, D. Vreugdenhil, M. Zabeau et al., 1996. Visualization of differential gene expression using a novel method of RNA fingerprinting based on AFLP: analysis of gene expression during potato tuber development. Plant J. 9: 745–753. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Miscellaneous

