Exploring the interaction of ruthenium(II) polypyridyl complexes with DNA using single-molecule techniques
- PMID: 16649785
- PMCID: PMC2519805
- DOI: 10.1021/la053242r
Exploring the interaction of ruthenium(II) polypyridyl complexes with DNA using single-molecule techniques
Abstract
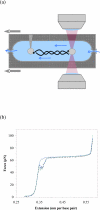
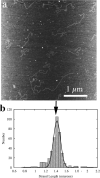
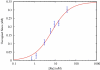
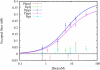
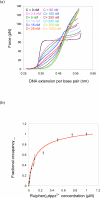
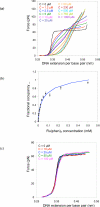
Here we explore DNA binding by a family of ruthenium(II) polypyridyl complexes using an atomic force microscope (AFM) and optical tweezers. We demonstrate using AFM that Ru(bpy)2dppz2+ intercalates into DNA (K(b) = 1.5 x 10(5) M(-1)), as does its close relative Ru(bpy)2dppx2+ (K(b) = 1.5 x 10(5) M(-1)). However, intercalation by Ru(phen)3(2+) and other Ru(II) complexes with K(b) values lower than that of Ru(bpy)2dppz2+ is difficult to determine using AFM because of competing aggregation and surface-binding phenomena. At the high Ru(II) concentrations required to evaluate intercalation, most of the DNA strands acquire a twisted, curled conformation that is impossible to measure accurately. The condensation of DNA on mica in the presence of polycations is well known, but it clearly precludes the accurate assessment by AFM of DNA intercalation by most Ru(II) complexes, though not by ethidium bromide and other monovalent intercalators. When stretching individual DNA molecules using optical tweezers, the same limitation on high metal concentration does not exist. Using optical tweezers, we show that Ru(phen)2dppz2+ intercalates avidly (K(b) = 3.2 x 10(6) M(-1)) whereas Ru(bpy)3(2+) does not intercalate, even at micromolar ruthenium concentrations. Ru(phen)3(2+) is shown to intercalate weakly (i.e., at micromolar concentrations (K(b) = 8.8 x 10(3) M(-1))). The distinct differences in DNA stretching behavior between Ru(phen)3(2+) and Ru(bpy)3(2+) clearly illustrate that intercalation can be distinguished from groove binding by pulling the DNA with optical tweezers. Our results demonstrate both the benefits and challenges of two single-molecule methods of exploring DNA binding and help to elucidate the mode of binding of Ru(phen)3(2+).
Figures



References
-
- Kumar CV, Barton JK, Turro NJ. J. Am. Chem. Soc. 1985;107:5518–5523.
-
- Barton JK, Goldberg JM, Kumar CV, Turro NJ. J. Am. Chem. Soc. 1986;108:2081–2088.
-
- Pyle AM, Rehmann JP, Meshoyrer R, Kumar CV, Turro NJ, Barton JK. J. Am. Chem. Soc. 1989;111:3051–3058.
-
- Hiort C, Nordén B, Rodger A. J. Am. Chem. Soc. 1990;112:1971–1982.
-
- Lincoln P, Nordén B. J. Phys. Chem. B. 1998;102:9583–9594.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

