Dynamic regulatory interactions of Polycomb group genes: MEDEA autoregulation is required for imprinted gene expression in Arabidopsis
- PMID: 16651654
- PMCID: PMC1472468
- DOI: 10.1101/gad.378106
Dynamic regulatory interactions of Polycomb group genes: MEDEA autoregulation is required for imprinted gene expression in Arabidopsis
Abstract
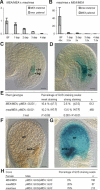
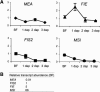
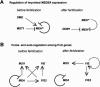
The imprinted Arabidopsis Polycomb group (PcG) gene MEDEA (MEA), which is homologous to Enhancer of Zeste [E(Z)], is maternally required for normal seed development. Here we show that, unlike known mammalian imprinted genes, MEA regulates its own imprinted expression: It down-regulates the maternal allele around fertilization and maintains the paternal allele silent later during seed development. Autorepression of the maternal MEA allele is direct and independent of the MEA-FIE (FERTILIZATION-INDEPENDENT ENDOSPERM) PcG complex, which is similar to the E(Z)-ESC (Extra sex combs) complex of animals, suggesting a novel mechanism. A complex network of cross-regulatory interactions among the other known members of the MEA-FIE PcG complex implies distinct functions that are dynamically regulated during reproduction.
Figures

References
-
- Brock H.W., van Lohuizen M. The Polycomb group: No longer an exclusive club? Curr. Opin. Genet. Dev. 2001;11:175–181. - PubMed
-
- Cao R., Zhang Y. The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr. Opin. Genet. Dev. 2004;14:155–164. - PubMed
-
- Chanvivattana Y., Bishopp A., Schubert D., Stock C., Moon Y.H., Sung Z.R., Goodrich J. Interaction of Polycomb-group proteins controlling flowering in Arabidopsis. Development. 2004;131:5263–5276. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
