A genome-wide distribution of 8-oxoguanine correlates with the preferred regions for recombination and single nucleotide polymorphism in the human genome
- PMID: 16651663
- PMCID: PMC1457041
- DOI: 10.1101/gr.4769606
A genome-wide distribution of 8-oxoguanine correlates with the preferred regions for recombination and single nucleotide polymorphism in the human genome
Abstract
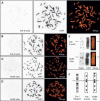
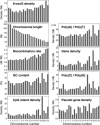
8-Oxoguanine (8-oxoG), a major spontaneous form of oxidative DNA damage, is considered to be a natural cause of genomic diversity in organisms because of its mutagenic potential. The steady-state level of 8-oxoG in the nuclear genome of a human cell has been estimated to be several residues per 10(6) guanines. In the present study, to clarify the genome-wide distribution of 8-oxoG in the steady state, we performed fluorescence in situ detection of 8-oxoG on human metaphase chromosomes using a monoclonal antibody. Multiple dot-like signals were observed on each metaphase chromosome. We then mapped the position of the signal at megabase resolution referring to the cytogenetically identified chromosomal band, and demonstrated that 8-oxoG is unevenly distributed in the normal human genome and that the distribution pattern is conserved among different individuals. Moreover, we found that regions with a high frequency of recombination and single nucleotide polymorphisms (SNPs) are preferentially located within chromosomal regions with a high density of 8-oxoG. Our findings suggest that 8-oxoG is one of the main causes of frequent recombinations and SNPs in the human genome, which largely contribute to the genomic diversity in human beings.
Figures



References
-
- Aburatani H., Hippo Y., Ishida T., Takashima R., Matsuba C., Kodama T., Takao M., Yasui A., Yamamoto K., Asano M., Hippo Y., Ishida T., Takashima R., Matsuba C., Kodama T., Takao M., Yasui A., Yamamoto K., Asano M., Ishida T., Takashima R., Matsuba C., Kodama T., Takao M., Yasui A., Yamamoto K., Asano M., Takashima R., Matsuba C., Kodama T., Takao M., Yasui A., Yamamoto K., Asano M., Matsuba C., Kodama T., Takao M., Yasui A., Yamamoto K., Asano M., Kodama T., Takao M., Yasui A., Yamamoto K., Asano M., Takao M., Yasui A., Yamamoto K., Asano M., Yasui A., Yamamoto K., Asano M., Yamamoto K., Asano M., Asano M. Cloning and characterization of mammalian 8-hydroxyguanine-specific DNA glycosylase/apurinic, apyrimidinic lyase, a functional mutM homologue. Cancer Res. 1997;57:2151–2156. - PubMed
-
- Aquadro C.F., Bauer Dumont V., Reed F.A., Bauer Dumont V., Reed F.A., Reed F.A. Genome-wide variation in the human and fruitfly: A comparison. Curr. Opin. Genet. Dev. 2001;11:627–634. - PubMed
-
- Arai T., Kelly V.P., Komoro K., Minowa O., Noda T., Nishimura S., Kelly V.P., Komoro K., Minowa O., Noda T., Nishimura S., Komoro K., Minowa O., Noda T., Nishimura S., Minowa O., Noda T., Nishimura S., Noda T., Nishimura S., Nishimura S. Cell proliferation in liver of Mmh/Ogg1-deficient mice enhances mutation frequency because of the presence of 8-hydroxyguanine in DNA. Cancer Res. 2003;63:4287–4292. - PubMed
-
- Barch M.J. The ACT cytogenetics laboratory manual. Raven Press; New York.: 1991.
-
- Barnes D.E., Lindahl T., Lindahl T. Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu. Rev. Genet. 2004;38:445–476. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
