The fate of laterally transferred genes: life in the fast lane to adaptation or death
- PMID: 16651664
- PMCID: PMC1457040
- DOI: 10.1101/gr.4746406
The fate of laterally transferred genes: life in the fast lane to adaptation or death
Abstract
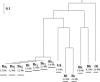
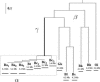
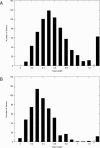
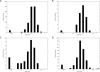
Large-scale genome arrangement plays an important role in bacterial genome evolution. A substantial number of genes can be inserted into, deleted from, or rearranged within genomes during evolution. Detecting or inferring gene insertions/deletions is of interest because such information provides insights into bacterial genome evolution and speciation. However, efficient inference of genome events is difficult because genome comparisons alone do not generally supply enough information to distinguish insertions, deletions, and other rearrangements. In this study, homologous genes from the complete genomes of 13 closely related bacteria were examined. The presence or absence of genes from each genome was cataloged, and a maximum likelihood method was used to infer insertion/deletion rates according to the phylogenetic history of the taxa. It was found that whole gene insertions/deletions in genomes occur at rates comparable to or greater than the rate of nucleotide substitution and that higher insertion/deletion rates are often inferred to be present at the tips of the phylogeny with lower rates on more ancient interior branches. Recently transferred genes are under faster and relaxed evolution compared with more ancient genes. Together, this implies that many of the lineage-specific insertions are lost quickly during evolution and that perhaps a few of the genes inserted by lateral transfer are niche specific.
Figures

Similar articles
-
The role of laterally transferred genes in adaptive evolution.BMC Evol Biol. 2007 Feb 8;7 Suppl 1(Suppl 1):S8. doi: 10.1186/1471-2148-7-S1-S8. BMC Evol Biol. 2007. PMID: 17288581 Free PMC article.
-
Uncovering rate variation of lateral gene transfer during bacterial genome evolution.BMC Genomics. 2008 May 20;9:235. doi: 10.1186/1471-2164-9-235. BMC Genomics. 2008. PMID: 18492275 Free PMC article.
-
Patterns of bacterial gene movement.Mol Biol Evol. 2004 Jul;21(7):1294-307. doi: 10.1093/molbev/msh129. Epub 2004 Apr 28. Mol Biol Evol. 2004. PMID: 15115802
-
Detecting lateral genetic transfer : a phylogenetic approach.Methods Mol Biol. 2008;452:457-69. doi: 10.1007/978-1-60327-159-2_21. Methods Mol Biol. 2008. PMID: 18566777 Review.
-
Lateral gene exchanges shape the genomes of amoeba-resisting microorganisms.Front Cell Infect Microbiol. 2012 Aug 21;2:110. doi: 10.3389/fcimb.2012.00110. eCollection 2012. Front Cell Infect Microbiol. 2012. PMID: 22919697 Free PMC article. Review.
Cited by
-
The ecology of bacterial genes and the survival of the new.Int J Evol Biol. 2012;2012:394026. doi: 10.1155/2012/394026. Epub 2012 Jul 31. Int J Evol Biol. 2012. PMID: 22900231 Free PMC article.
-
Horizontal gene transfer and the evolution of transcriptional regulation in Escherichia coli.Genome Biol. 2008 Jan 7;9(1):R4. doi: 10.1186/gb-2008-9-1-r4. Genome Biol. 2008. PMID: 18179685 Free PMC article.
-
Horizontal transfer and gene conversion as an important driving force in shaping the landscape of mitochondrial introns.G3 (Bethesda). 2014 Apr 16;4(4):605-12. doi: 10.1534/g3.113.009910. G3 (Bethesda). 2014. PMID: 24515269 Free PMC article.
-
Inference of gain and loss events from phyletic patterns using stochastic mapping and maximum parsimony--a simulation study.Genome Biol Evol. 2011;3:1265-75. doi: 10.1093/gbe/evr101. Epub 2011 Oct 4. Genome Biol Evol. 2011. PMID: 21971516 Free PMC article.
-
DiscML: an R package for estimating evolutionary rates of discrete characters using maximum likelihood.BMC Bioinformatics. 2014 Sep 27;15(1):320. doi: 10.1186/1471-2105-15-320. BMC Bioinformatics. 2014. PMID: 25260628 Free PMC article.
References
-
- Ash C., Farrow J.A., Dorsch M., Stackebrandt E., Collins M.D., Farrow J.A., Dorsch M., Stackebrandt E., Collins M.D., Dorsch M., Stackebrandt E., Collins M.D., Stackebrandt E., Collins M.D., Collins M.D. Comparative analysis of Bacillus anthracis, Bacillus cereus, and related species on the basis of reverse transcriptase sequencing of 16S rRNA. Int. J. Syst. Bacteriol. 1991;41:343–346. - PubMed
-
- Brunder W., Karch H., Karch H. Genome plasticity in Enterobacteriaceae. Int. J. Med. Microbiol. 2000;290:153–165. - PubMed
-
- Cerdeno-Tarraga A.M., Patrick S., Crossman L.C., Blakely G., Abratt V., Lennard N., Poxton I., Duerden B., Harris B., Quail M.A., Patrick S., Crossman L.C., Blakely G., Abratt V., Lennard N., Poxton I., Duerden B., Harris B., Quail M.A., Crossman L.C., Blakely G., Abratt V., Lennard N., Poxton I., Duerden B., Harris B., Quail M.A., Blakely G., Abratt V., Lennard N., Poxton I., Duerden B., Harris B., Quail M.A., Abratt V., Lennard N., Poxton I., Duerden B., Harris B., Quail M.A., Lennard N., Poxton I., Duerden B., Harris B., Quail M.A., Poxton I., Duerden B., Harris B., Quail M.A., Duerden B., Harris B., Quail M.A., Harris B., Quail M.A., Quail M.A., et al. Extensive DNA inversions in the B. fragilis genome control variable gene expression. Science. 2005;307:1463–1465. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
