Minimization and optimization of designed beta-hairpin folds
- PMID: 16669679
- PMCID: PMC3164952
- DOI: 10.1021/ja054971w
Minimization and optimization of designed beta-hairpin folds
Abstract
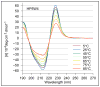
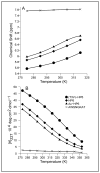
Minimized beta hairpins have provided additional data on the geometric preferences of Trp interactions in TW-loop-WT motifs. This motif imparts significant fold stability to peptides as short as 8 residues. High-resolution NMR structures of a 16- (KKWTWNPATGKWTWQE, DeltaG(U)(298) >or= +7 kJ/mol) and 12-residue (KTWNPATGKWTE, DeltaG(U)(298) = +5.05 kJ/mol) hairpin reveal a common turn geometry and edge-to-face (EtF) packing motif and a cation-pi interaction between Lys(1) and the Trp residue nearest the C-terminus. The magnitude of a CD exciton couplet (due to the two Trp residues) and the chemical shifts of a Trp Hepsilon3 site (shifted upfield by 2.4 ppm due to the EtF stacking geometry) provided near-identical measures of folding. CD melts of representative peptides with the -TW-loop-WT- motif provided the thermodynamic parameters for folding, which reflect enthalpically driven folding at laboratory temperatures with a small DeltaC(p) for unfolding (+420 J K(-)(1)/mol). In the case of Asx-Pro-Xaa-Thr-Gly-Xaa loops, mutations established that the two most important residues in this class of direction-reversing loops are Asx and Gly: mutation to alanine is destabilizing by about 6 and 2 kJ/mol, respectively. All indicators of structuring are retained in a minimized 8-residue construct (Ac-WNPATGKW-NH(2)) with the fold stability reduced to DeltaG(U)(278) = -0.7 kJ/mol. NMR and CD comparisons indicate that -TWXNGKWT- (X = S, I) sequences also form the same hairpin-stabilizing W/W interaction.
Figures




References
-
- Ramírez-Alvarado M, Blanco FJ, Serrano L. Nat Struct Biol. 1996;3:604–612. - PubMed
- Maynard AJ, Sharman GJ, Searle MS. J Am Chem Soc. 1998;120:1996–2007.
- Stanger HE, Gellman SH. J Am Chem Soc. 1998;120:4236–4237.
- Syud FA, Espinosa JF, Gellman SH. J Am Chem Soc. 1999;121:11577–11578.
-
- Griffiths-Jones SR, Maynard AJ, Searle MS. J Mol Biol. 1999;292:1051–1069. - PubMed
-
- de Alba E, Jiménez MA, Rico M. J Am Chem Soc. 1997;119:175–183.
-
- Fesinmeyer RM, Hudson FM, Andersen NH. J Am Chem Soc. 2004;126:7238–7243. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

