Functional interactions of the SPAK/OSR1 kinases with their upstream activator WNK1 and downstream substrate NKCC1
- PMID: 16669787
- PMCID: PMC1479760
- DOI: 10.1042/BJ20060220
Functional interactions of the SPAK/OSR1 kinases with their upstream activator WNK1 and downstream substrate NKCC1
Abstract
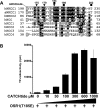
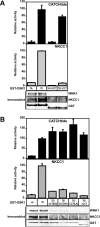
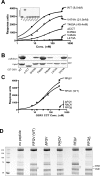
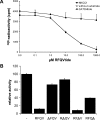
The SPAK (STE20/SPS1-related proline/alanine-rich kinase) and OSR1 (oxidative stress-responsive kinase-1) kinases interact and phosphorylate NKCC1 (Na+-K+-2Cl- co-transporter-1), leading to its activation. Recent studies indicated that SPAK and OSR1 are phosphorylated and activated by the WNK1 [with no K (lysine) protein kinase-1] and WNK4, genes mutated in humans affected by Gordon's hypertension syndrome. In the present study, we have identified three residues in NKCC1 (Thr175/Thr179/Thr184 in shark or Thr203/Thr207/Thr212 in human) that are phosphorylated by SPAK and OSR1, and have developed a peptide substrate, CATCHtide (cation chloride co-transporter peptide substrate), to assess SPAK and OSR1 activity. Exposure of HEK-293 (human embryonic kidney) cells to osmotic stress, which leads to phosphorylation and activation of NKCC1, increased phosphorylation of NKCC1 at the sites targeted by SPAK/OSR1. The residues on NKCC1, phosphorylated by SPAK/OSR1, are conserved in other cation co-transporters, such as the Na+-Cl- co-transporter, the target of thiazide drugs that lower blood pressure in humans with Gordon's syndrome. Furthermore, we characterize the properties of a 92-residue CCT (conserved C-terminal) domain on SPAK and OSR1 that interacts with an RFXV (Arg-Phe-Xaa-Val) motif present in the substrate NKCC1 and its activators WNK1/WNK4. A peptide containing the RFXV motif interacts with nanomolar affinity with the CCT domains of SPAK/OSR1 and can be utilized to affinity-purify SPAK and OSR1 from cell extracts. Mutation of the arginine, phenylalanine or valine residue within this peptide abolishes binding to SPAK/OSR1. We have identified specific residues within the CCT domain that are required for interaction with the RFXV motif and have demonstrated that mutation of these in OSR1 inhibited phosphorylation of NKCC1, but not of CATCHtide which does not possess an RFXV motif. We establish that an intact CCT domain is required for WNK1 to efficiently phosphorylate and activate OSR1. These data establish that the CCT domain functions as a multipurpose docking site, enabling SPAK/OSR1 to interact with substrates (NKCC1) and activators (WNK1/WNK4).
Figures


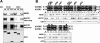

References
-
- Piechotta K., Lu J., Delpire E. Cation chloride cotransporters interact with the stress-related kinases Ste20-related proline-alanine-rich kinase (SPAK) and oxidative stress response 1 (OSR1) J. Biol. Chem. 2002;277:50812–50819. - PubMed
-
- Dowd B. F., Forbush B. PASK (proline-alanine-rich STE20-related kinase), a regulatory kinase of the Na–K–Cl cotransporter (NKCC1) J. Biol. Chem. 2003;278:27347–27353. - PubMed
-
- Gagnon K. B., England R., Delpire E. Volume sensitivity of cation–Cl− cotransporters is modulated by the interaction of two kinases: Ste20-related prolinealanine-rich kinase and WNK4. Am. J. Physiol. Cell Physiol. 2006;290:C134–C142. - PubMed
-
- Piechotta K., Garbarini N., England R., Delpire E. Characterization of the interaction of the stress kinase SPAK with the Na+–K+–2Cl− cotransporter in the nervous system: evidence for a scaffolding role of the kinase. J. Biol. Chem. 2003;278:52848–52856. - PubMed
-
- Moriguchi T., Urushiyama S., Hisamoto N., Iemura S., Uchida S., Natsume T., Matsumoto K., Shibuya H. WNK1 regulates phosphorylation of cation–chloride-coupled cotransporters via the STE20-related kinases, SPAK and OSR1. J. Biol. Chem. 2005;280:42685–42693. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

