Human LINE-1 retrotransposon induces DNA damage and apoptosis in cancer cells
- PMID: 16670018
- PMCID: PMC1464142
- DOI: 10.1186/1475-2867-6-13
Human LINE-1 retrotransposon induces DNA damage and apoptosis in cancer cells
Abstract
Background: Long interspersed nuclear elements (LINEs), Alu and endogenous retroviruses (ERVs) make up some 45% of human DNA. LINE-1 also called L1, is the most common family of non-LTR retrotransposons in the human genome and comprises about 17% of the genome. L1 elements require the integration into chromosomal target sites using L1-encoded endonuclease which creates staggering DNA breaks allowing the newly transposed L1 copies to integrate into the genome. L1 expression and retrotransposition in cancer cells might cause transcriptional deregulation, insertional mutations, DNA breaks, and an increased frequency of recombinations, contributing to genome instability. There is however little evidence on the mechanism of L1-induced genetic instability and its impact on cancer cell growth and proliferation.
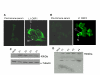
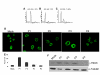
Results: We report that L1 has genome-destabilizing effects indicated by an accumulation of gamma-H2AX foci, an early response to DNA strand breaks, in association with an abnormal cell cycle progression through a G2/M accumulation and an induction of apoptosis in breast cancer cells. In addition, we found that adjuvant L1 activation may lead to supra-additive killing when combined with radiation by enhancing the radiation lethality through induction of apoptosis that we have detected through Bax activation.
Conclusion: L1 retrotransposition is sensed as a DNA damaging event through the creation DNA breaks involving L1-encoded endonuclease. The apparent synergistic interaction between L1 activation and radiation can further be utilized for targeted induction of cancer cell death. Thus, the role of retrotransoposons in general, and of L1 in particular, in DNA damage and repair assumes larger significance both for the understanding of mutagenicity and, potentially, for the control of cell proliferation and apoptosis.
Figures



References
-
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann N, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, Gibbs RA, Muzny DM, Scherer SE, Bouck JB, Sodergren EJ, Worley KC, Rives CM, Gorrell JH, Metzker ML, Naylor SL, Kucherlapati RS, Nelson DL, Weinstock GM, Sakaki Y, Fujiyama A, Hattori M, Yada T, Toyoda A, Itoh T, Kawagoe C, Watanabe H, Totoki Y, Taylor T, Weissenbach J, Heilig R, Saurin W, Artiguenave F, Brottier P, Bruls T, Pelletier E, Robert C, Wincker P, Smith DR, Doucette-Stamm L, Rubenfield M, Weinstock K, Lee HM, Dubois J, Rosenthal A, Platzer M, Nyakatura G, Taudien S, Rump A, Yang H, Yu J, Wang J, Huang G, Gu J, Hood L, Rowen L, Madan A, Qin S, Davis RW, Federspiel NA, Abola AP, Proctor MJ, Myers RM, Schmutz J, Dickson M, Grimwood J, Cox DR, Olson MV, Kaul R, Raymond C, Shimizu N, Kawasaki K, Minoshima S, Evans GA, Athanasiou M, Schultz R, Roe BA, Chen F, Pan H, Ramser J, Lehrach H, Reinhardt R, McCombie WR, de la Bastide M, Dedhia N, Blocker H, Hornischer K, Nordsiek G, Agarwala R, Aravind L, Bailey JA, Bateman A, Batzoglou S, Birney E, Bork P, Brown DG, Burge CB, Cerutti L, Chen HC, Church D, Clamp M, Copley RR, Doerks T, Eddy SR, Eichler EE, Furey TS, Galagan J, Gilbert JG, Harmon C, Hayashizaki Y, Haussler D, Hermjakob H, Hokamp K, Jang W, Johnson LS, Jones TA, Kasif S, Kaspryzk A, Kennedy S, Kent WJ, Kitts P, Koonin EV, Korf I, Kulp D, Lancet D, Lowe TM, McLysaght A, Mikkelsen T, Moran JV, Mulder N, Pollara VJ, Ponting CP, Schuler G, Schultz J, Slater G, Smit AF, Stupka E, Szustakowski J, Thierry-Mieg D, Thierry-Mieg J, Wagner L, Wallis J, Wheeler R, Williams A, Wolf YI, Wolfe KH, Yang SP, Yeh RF, Collins F, Guyer MS, Peterson J, Felsenfeld A, Wetterstrand KA, Patrinos A, Morgan MJ, de Jong P, Catanese JJ, Osoegawa K, Shizuya H, Choi S, Chen YJL. International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

