Homologous recombination prevents methylation-induced toxicity in Escherichia coli
- PMID: 16670432
- PMCID: PMC1456334
- DOI: 10.1093/nar/gkl222
Homologous recombination prevents methylation-induced toxicity in Escherichia coli
Abstract
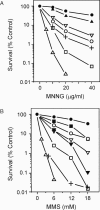
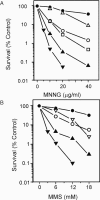
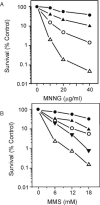
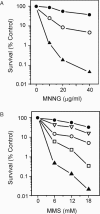
Methylating agents such as N-methyl-N'-nitro-N-nitrosoguanidine (MNNG) and methyl methane sulfonate (MMS) produce a wide variety of N- and O-methylated bases in DNA, some of which can block replication fork progression. Homologous recombination is a mechanism by which chromosome replication can proceed despite the presence of lesions. The two major recombination pathways, RecBCD and RecFOR, which repair double-strand breaks (DSBs) and single-strand gaps respectively, are needed to protect against toxicity with the RecBCD system being more important. We find that recombination-deficient cell lines, such as recBCD recF, and ruvC recG, are as sensitive to the cytotoxic effects of MMS and MNNG as the most base excision repair (BER)-deficient (alkA tag) isogenic mutant strain. Recombination and BER-deficient double mutants (alkA tag recBCD) were more sensitive to MNNG and MMS than the single mutants suggesting that homologous recombination and BER play essential independent roles. Cells deleted for the polA (DNA polymerase I) or priA (primosome) genes are as sensitive to MMS and MNNG as alkA tag bacteria. Our results suggest that the mechanism of cytotoxicity by alkylating agents includes the necessity for homologous recombination to repair DSBs and single-strand gaps produced by DNA replication at blocking lesions or single-strand nicks resulting from AP-endonuclease action.
Figures



References
-
- Sedgwick B. Repairing DNA-methylation damage. Nat. Rev. Mol. Cell Biol. 2004;5:148–157. - PubMed
-
- Strauss B., Scudiero D., Henderson E. The nature of the alkylation lesion in mammalian cells. Basic Life Sci. 1975;5A:13–24. - PubMed
-
- Boiteux S., Laval J. Mutagenesis by alkylating agents: coding properties for DNA polymerase of poly (dC) template containing 3-methylcytosine. Biochimie. 1982;64:637–641. - PubMed
-
- Larson K., Sahm J., Shenkar R., Strauss B. Methylation-induced blocks to in vitro DNA replication. Mutat. Res. 1985;150:77–84. - PubMed

