Mobile D-loops are a preferred substrate for the Bloom's syndrome helicase
- PMID: 16670433
- PMCID: PMC1456333
- DOI: 10.1093/nar/gkl258
Mobile D-loops are a preferred substrate for the Bloom's syndrome helicase
Abstract
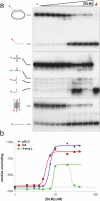
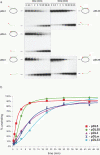
The Bloom's syndrome helicase, BLM, is a member of the highly conserved RecQ family, and possesses both DNA unwinding and DNA strand annealing activities. BLM also promotes branch migration of Holliday junctions. One role for BLM is to act in conjunction with topoisomerase IIIalpha to process homologous recombination (HR) intermediates containing a double Holliday junction by a process termed dissolution. However, several lines of evidence suggest that BLM may also act early in one or more of the recombination pathways to eliminate illegitimate or aberrantly paired DNA joint molecules. We have investigated whether BLM can disrupt DNA displacement loops (D-loops), which represent the initial strand invasion step of HR. We show that mobile D-loops created by the RecA recombinase are a highly preferred substrate for BLM with the invading strand being displaced from the duplex. We have identified structural features of the D-loop that determine the efficiency with which BLM promotes D-loop dissociation. We discuss these results in the context of models for the role of BLM as an 'anti-recombinase'.
Figures




References
-
- German J. Bloom syndrome: a mendelian prototype of somatic mutational disease. Medicine (Baltimore) 1993;72:393–406. - PubMed
-
- Ellis N.A., Groden J., Ye T.Z., Straughen J., Lennon D.J., Ciocci S., Proytcheva M., German J. The Bloom's syndrome gene product is homologous to RecQ helicases. Cell. 1995;83:655–666. - PubMed
-
- Karow J.K., Chakraverty R.K., Hickson I.D. The Bloom's syndrome gene product is a 3′–5′ DNA helicase. J. Biol. Chem. 1997;272:30611–30614. - PubMed

