Four genomic islands that mark post-1995 pandemic Vibrio parahaemolyticus isolates
- PMID: 16672049
- PMCID: PMC1464126
- DOI: 10.1186/1471-2164-7-104
Four genomic islands that mark post-1995 pandemic Vibrio parahaemolyticus isolates
Abstract
Background: Vibrio parahaemolyticus is an aquatic, halophilic, Gram-negative bacterium, first discovered in 1950 in Japan during a food-poisoning outbreak. Infections resulting from consumption of V. parahaemolyticus have increased globally in the last 10 years leading to the bacterium's classification as a newly emerging pathogen. In 1996 the first appearance of a pandemic V. parahaemolyticus clone occurred, a new O3:K6 serotype strain that has now been identified worldwide as a major cause of seafood-borne gastroenteritis.
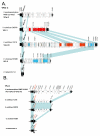
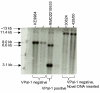
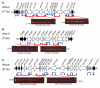
Results: We examined the sequenced genome of V. parahaemolyticus RIMD2210633, an O3:K6 serotype strain isolated in Japan in 1996, by bioinformatic analyses to uncover genomic islands (GIs) that may play a role in the emergence and pathogenesis of pandemic strains. We identified 7 regions ranging in size from 10 kb to 81 kb that had the characteristics of GIs such as aberrant base composition compared to the core genome, presence of phage-like integrases, flanked by direct repeats and the absence of these regions from closely related species. Molecular analysis of worldwide clinical isolates of V. parahaemolyticus recovered over the last 33 years demonstrated that a 24 kb region named V. parahaemolyticus island-1 (VPaI-1) encompassing ORFs VP0380 to VP0403 is only present in new O3:K6 and related strains recovered after 1995. We investigated the presence of 3 additional regions, VPaI-4 (VP2131 to VP2144), VPaI-5 (VP2900 to VP2910) and VPaI-6 (VPA1254 to VPA1270) by PCR assays and Southern blot analyses among the same set of V. parahaemolyticus isolates. These 3 VPaI regions also gave similar distribution patterns amongst the 41 strains examined.
Conclusion: The 4 VPaI regions examined may represent DNA acquired by the pandemic group of V. parahaemolyticus isolates that increased their fitness either in the aquatic environment or in their ability to infect humans.
Figures




References
-
- Okuda J, Ishibashi M, Hayakawa E, Nishino T, Takeda Y, Mukhopadhyay AK, Garg S, Bhattacharya SK, Nair GB, Nishibuchi M. Emergence of a unique O3:K6 clone of Vibrio parahaemolyticus in Calcutta, India, and isolation of strains from the same clonal group from Southeast Asian travelers arriving in Japan. J Clin Microbiol. 1997;35:3150–3155. - PMC - PubMed
-
- Abbott SL, Powers C, Kaysner CA, Takeda Y, Ishibashi M, Joseph SW, Janda JM. Emergence of a restricted bioserovar of Vibrio parahaemolyticus as the predominant cause of Vibrio-associated gastroenteritis on the West Coast of the United States and Mexico. J Clin Microbiol. 1989;27:2891–2893. - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

