Structural characterization of GntR/HutC family signaling domain
- PMID: 16672238
- PMCID: PMC2242532
- DOI: 10.1110/ps.062146906
Structural characterization of GntR/HutC family signaling domain
Abstract
The crystal structure of Escherichia coli PhnF C-terminal domain (C-PhnF) was solved at 1.7 A resolution by the single wavelength anomalous dispersion (SAD) method. The PhnF protein belongs to the HutC subfamily of the large GntR transcriptional regulator family. Members of this family share similar N-terminal DNA-binding domains, but are divided into four subfamilies according to their heterogenic C-terminal domains, which are involved in effector binding and oligomerization. The C-PhnF structure provides for the first time the scaffold of this domain for the HutC subfamily, which covers about 31% of GntR-like regulators. The structure represents a mixture of alpha-helices and beta-strands, with a six-stranded antiparallel beta-sheet at the core. C-PhnF monomers form a dimer by establishing interdomain eight-strand beta-sheets that include core antiparallel and N-terminal two-strand parallel beta-sheets from each monomer. C-PhnF shares strong structural similarity with the chorismate lyase fold, which features a buried active site locked behind two helix-turn-helix loops. The structural comparison of the C-PhnF and UbiC proteins allows us to propose that a similar site in the PhnF structure is adapted for effector binding.
Figures

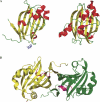

Similar articles
-
Insight into the induction mechanism of the GntR/HutC bacterial transcription regulator YvoA.Nucleic Acids Res. 2010 Apr;38(7):2485-97. doi: 10.1093/nar/gkp1191. Epub 2010 Jan 4. Nucleic Acids Res. 2010. PMID: 20047956 Free PMC article.
-
HutC/FarR-like bacterial transcription factors of the GntR family contain a small molecule-binding domain of the chorismate lyase fold.FEMS Microbiol Lett. 2003 May 16;222(1):17-23. doi: 10.1016/S0378-1097(03)00242-8. FEMS Microbiol Lett. 2003. PMID: 12757941
-
Subdivision of the helix-turn-helix GntR family of bacterial regulators in the FadR, HutC, MocR, and YtrA subfamilies.J Biol Chem. 2002 Apr 12;277(15):12507-15. doi: 10.1074/jbc.M110968200. Epub 2001 Dec 27. J Biol Chem. 2002. PMID: 11756427
-
Allosteric control of transcription in GntR family of transcription regulators: A structural overview.IUBMB Life. 2015 Jul;67(7):556-63. doi: 10.1002/iub.1401. Epub 2015 Jul 14. IUBMB Life. 2015. PMID: 26172911 Review.
-
Members of the IclR family of bacterial transcriptional regulators function as activators and/or repressors.FEMS Microbiol Rev. 2006 Mar;30(2):157-86. doi: 10.1111/j.1574-6976.2005.00008.x. FEMS Microbiol Rev. 2006. PMID: 16472303 Review.
Cited by
-
A Salmonella Regulator Modulates Intestinal Colonization and Use of Phosphonoacetic Acid.Front Cell Infect Microbiol. 2017 Mar 15;7:69. doi: 10.3389/fcimb.2017.00069. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 28361036 Free PMC article.
-
Insight into the induction mechanism of the GntR/HutC bacterial transcription regulator YvoA.Nucleic Acids Res. 2010 Apr;38(7):2485-97. doi: 10.1093/nar/gkp1191. Epub 2010 Jan 4. Nucleic Acids Res. 2010. PMID: 20047956 Free PMC article.
-
Global Regulatory Roles of the Histidine-Responsive Transcriptional Repressor HutC in Pseudomonas fluorescens SBW25.J Bacteriol. 2020 Jun 9;202(13):e00792-19. doi: 10.1128/JB.00792-19. Print 2020 Jun 9. J Bacteriol. 2020. PMID: 32291279 Free PMC article.
-
Structural and functional characterization of the LldR from Corynebacterium glutamicum: a transcriptional repressor involved in L-lactate and sugar utilization.Nucleic Acids Res. 2008 Dec;36(22):7110-23. doi: 10.1093/nar/gkn827. Epub 2008 Nov 6. Nucleic Acids Res. 2008. PMID: 18988622 Free PMC article.
-
A genomic approach to understand interactions between Streptococcus pneumoniae and its bacteriophages.BMC Genomics. 2015 Nov 18;16:972. doi: 10.1186/s12864-015-2134-8. BMC Genomics. 2015. PMID: 26582495 Free PMC article.
References
-
- Aravind L. and Anantharaman V. 2003. HutC/FarR-like bacterial transcription factors of the GntR family contain a small molecule-binding domain of the chorismate lyase fold FEMS Microbiol. Lett. 222 17–23. - PubMed
-
- Gallagher D.T., Mayhew M., Holden M.J., Howard A., Kim K.J., Vilker V.L. 2001. The crystal structure of chorismate lyase shows a new fold and a tightly retained product Proteins 44 304–311. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

