Elimination of damaged proteins during differentiation of embryonic stem cells
- PMID: 16672370
- PMCID: PMC1472508
- DOI: 10.1073/pnas.0510944103
Elimination of damaged proteins during differentiation of embryonic stem cells
Abstract
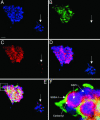
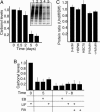
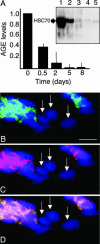
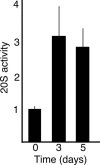
During mammalian aging, cellular proteins become increasingly damaged: for example, by carbonylation and formation of advanced glycation end products (AGEs). The means to ensure that offspring are born without such damage are unknown. Unexpectedly, we found that undifferentiated mouse ES cells contain high levels of both carbonyls and AGEs. The damaged proteins, identified as chaperones and proteins of the cytoskeleton, are the main targets for protein oxidation in aged tissues. However, the mouse ES cells rid themselves of such damage upon differentiation in vitro. This elimination of damaged proteins coincides with a considerably elevated activity of the 20S proteasome. Moreover, damaged proteins were primarily observed in the inner cell mass of blastocysts, whereas the cells that had embarked on differentiation into the trophectoderm displayed drastically reduced levels of protein damage. Thus, the elimination of protein damage occurs also during normal embryonic development in vivo. This clear-out of damaged proteins may be a part of a previously unknown rejuvenation process at the protein level that occurs at a distinct stage during early embryonic development.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures

References
-
- Lombard D. B., Chua K. F., Mostoslavsky R., Franco S., Gostissa M., Alt F. W. Cell. 2005;120:497–512. - PubMed
-
- Medvedev Z. A. Mech. Ageing Dev. 1981;4:331–359. - PubMed
-
- Stadtman E. R. Science. 1992;257:1220–1224. - PubMed
-
- Levine R. L. Free Radical Biol. Med. 2002;32:790–796. - PubMed
-
- Bota D. A., Davies K. J. Nat. Cell Biol. 2002;4:674–680. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

