Diversity and distribution of marine microbial eukaryotes in the Arctic Ocean and adjacent seas
- PMID: 16672445
- PMCID: PMC1472370
- DOI: 10.1128/AEM.72.5.3085-3095.2006
Diversity and distribution of marine microbial eukaryotes in the Arctic Ocean and adjacent seas
Abstract
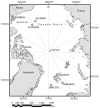
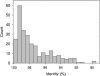
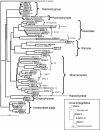
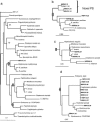
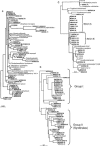
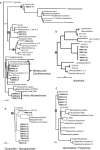
We analyzed microbial eukaryote diversity in perennially cold arctic marine waters by using 18S rRNA gene clone libraries. Samples were collected during concurrent oceanographic missions to opposite sides of the Arctic Ocean Basin and encompassed five distinct water masses. Two deep water Arctic Ocean sites and the convergence of the Greenland, Norwegian, and Barents Seas were sampled from 28 August to 2 September 2002. An additional sample was obtained from the Beaufort Sea (Canada) in early October 2002. The ribotypes were diverse, with different communities among sites and between the upper mixed layer and just below the halocline. Eukaryotes from the remote Canada Basin contained new phylotypes belonging to the radiolarian orders Acantharea, Polycystinea, and Taxopodida. A novel group within the photosynthetic stramenopiles was also identified. One sample closest to the interior of the Canada Basin yielded only four major taxa, and all but two of the sequences recovered belonged to the polar diatom Fragilariopsis and a radiolarian. Overall, 42% of the sequences were <98% similar to any sequences in GenBank. Moreover, 15% of these were <95% similar to previously recovered sequences, which is indicative of endemic or undersampled taxa in the North Polar environment. The cold, stable Arctic Ocean is a threatened environment, and climate change could result in significant loss of global microbial biodiversity.
Figures
References
-
- Aagaard, K., L. K. Coachman, and E. Carmack. 1981. On the halocline of the Arctic Ocean. Deep-Sea Res. A 28:529.
-
- ACIA. 2005. Arctic climate impact assessment. Cambridge University Press, Cambridge, United Kingdom.
-
- Afanasieva, M. S., and E. O. Amon. 2003. A new classification of the Radiolaria. Paleontol. J. 37:630-645.
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Meyers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403. - PubMed
-
- Baldauf, S. L. 2003. The deep roots of eukaryotes. Science 300:1703-1706. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

