The ornibactin biosynthesis and transport genes of Burkholderia cenocepacia are regulated by an extracytoplasmic function sigma factor which is a part of the Fur regulon
- PMID: 16672617
- PMCID: PMC1482860
- DOI: 10.1128/JB.188.10.3631-3644.2006
The ornibactin biosynthesis and transport genes of Burkholderia cenocepacia are regulated by an extracytoplasmic function sigma factor which is a part of the Fur regulon
Abstract
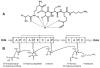
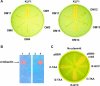
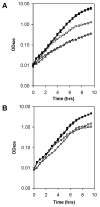
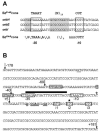
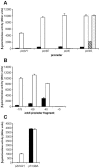
Burkholderia cenocepacia mutants that fail to produce the siderophore ornibactin were obtained following mutagenesis with mini-Tn5Tp. These mutants were shown to be growth restricted under conditions of iron depletion. In eight of the mutants, the transposon had integrated into one of two genes, orbI and orbJ, encoding nonribosomal peptide synthetases. In the other mutant, the transposon had inserted into an open reading frame, orbS, located upstream from orbI. The polypeptide product of orbS exhibits a high degree of similarity to the Pseudomonas aeruginosa extracytoplasmic function (ECF) sigma factor PvdS but possesses an N-terminal extension of approximately 29 amino acids that is not present in PvdS. Three predicted OrbS-dependent promoters were identified within the ornibactin gene cluster, based on their similarity to PvdS-dependent promoters. The iron-regulated activity of these promoters was shown to require OrbS. Transcription of the orbS gene was found to be under the control of an iron-regulated sigma(70)-dependent promoter. This promoter, but not the OrbS-dependent promoters, was shown to be a target for repression by the global regulator Fur. Our results demonstrate that production of ornibactin by B. cenocepacia in response to iron starvation requires transcription of an operon that is dependent on the Fur-regulated ECF sigma factor gene orbS. A mechanism is also proposed for the biosynthesis of ornibactin.
Figures


References
-
- Aaron, S. D., W. Ferris, D. A. Henry, D. P. Speert, and N. E. Macdonald. 2000. Multiple combination bactericidal antibiotic testing for patients with cystic fibrosis infected with Burkholderia cepacia. Am. J. Respir. Crit. Care Med. 161:1206-1212. - PubMed
-
- Alexeyev, M. F. 1999. The pKNOCK series of broad-host-range mobilisable suicide vectors for gene knockout and targeted DNA insertion into the chromosome of gram-negative bacteria. BioTechniques 26:824-828. - PubMed
-
- Amman, E., B. Ochs, and K.-J. Abel. 1988. Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene 69:301-315. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

