Identification of a mutation in the Bacillus subtilis S-adenosylmethionine synthetase gene that results in derepression of S-box gene expression
- PMID: 16672621
- PMCID: PMC1482843
- DOI: 10.1128/JB.188.10.3674-3681.2006
Identification of a mutation in the Bacillus subtilis S-adenosylmethionine synthetase gene that results in derepression of S-box gene expression
Abstract
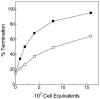
Genes in the S-box family are regulated by binding of S-adenosylmethionine (SAM) to the 5' region of the mRNA of the regulated gene. SAM binding was previously shown to promote a rearrangement of the RNA structure that results in premature termination of transcription in vitro and repression of expression of the downstream coding sequence. The S-box RNA element therefore acts as a SAM-binding riboswitch in vitro. In an effort to identify factors other than SAM that could be involved in the S-box regulatory mechanism in vivo, we searched for trans-acting mutations in Bacillus subtilis that act to disrupt repression of S-box gene expression during growth under conditions where SAM pools are elevated. We identified a single mutant that proved to have one nucleotide substitution in the metK gene, encoding SAM synthetase. This mutation, designated metK10, resulted in a 15-fold decrease in SAM synthetase activity and a 4-fold decrease in SAM concentration in vivo. The metK10 mutation specifically affected S-box gene expression, and the increase in expression under repressing conditions was dependent on the presence of a functional transcriptional antiterminator element. The observation that the mutation identified in this search affects SAM production supports the model that the S-box RNAs directly monitor SAM in vivo, without a requirement for additional factors.
Figures
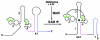

References
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
-
- Fuchs, R. T., F. J. Grundy, and T. M. Henkin. 2006. The SMK box is a new SAM-binding RNA for translational regulation of SAM synthetase. Nat. Struct. Mol. Biol. 13:226-233. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

