Reduction/oxidation-phosphorylation control of DNA binding in the bZIP dimerization network
- PMID: 16674813
- PMCID: PMC1479340
- DOI: 10.1186/1471-2164-7-107
Reduction/oxidation-phosphorylation control of DNA binding in the bZIP dimerization network
Abstract
Background: bZIPs are transcription factors that are found throughout the eukarya from fungi to flowering plants and mammals. They contain highly conserved basic region (BR) and leucine zipper (LZ) domains and often function as environmental sensors. Specifically, bZIPs frequently have a role in mediating the response to oxidative stress, a crucial environmental signal that needs to be transduced to the gene regulatory network.
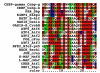
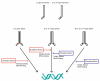
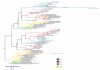
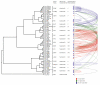
Results: Based on sequence comparisons and experimental data on a number of important bZIP transcription factors, we predict which bZIPs are under redox control and which are regulated via protein phosphorylation. By integrating genomic, phylogenetic and functional data from the literature, we then propose a link between oxidative stress and the choice of interaction partners for the bZIP proteins.
Conclusion: This integration permits the bZIP dimerization network to be interpreted in functional terms, especially in the context of the role of bZIP proteins in the response to environmental stress. This analysis demonstrates the importance of abiotic factors in shaping regulatory networks.
Figures

References
-
- Iguchi-Ariga SM, Schaffner W. CpG methylation of the cAMP-responsive enhancer/promoter sequence TGACGTCA abolishes specific factor binding as well as transcriptional activation. Genes Dev. 1989;3:612–9. - PubMed
-
- Mahoney CW, Shuman J, McKnight SL, Chen HC, Huang KP. Phosphorylation of CCAAT-enhancer binding protein by protein kinase C attenuates site-selective DNA binding. J Biol Chem. 1992;267:19396–403. - PubMed
-
- Abate C, Patel L, Rauscher FJ, 3rd, Curran T. Redox regulation of fos and jun DNA-binding activity in vitro. Science. 1990;249:1157–61. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

