Crystal structure of the SOCS2-elongin C-elongin B complex defines a prototypical SOCS box ubiquitin ligase
- PMID: 16675548
- PMCID: PMC1472497
- DOI: 10.1073/pnas.0601638103
Crystal structure of the SOCS2-elongin C-elongin B complex defines a prototypical SOCS box ubiquitin ligase
Abstract
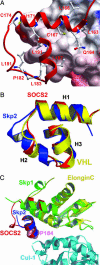
Growth hormone (GH) signaling is tightly controlled by ubiquitination of GH receptors, phosphorylation levels, and accessibility of binding sites for downstream signaling partners. Members of the suppressors of cytokine signaling (SOCS) family function as key regulators at all levels of this pathway, and mouse knockout studies implicate SOCS2 as the primary suppressor. To elucidate the structural basis for SOCS2 function, we determined the 1.9-A crystal structure of the ternary complex of SOCS2 with elongin C and elongin B. The structure defines a prototypical SOCS box ubiquitin ligase with a Src homology 2 (SH2) domain as a substrate recognition motif. Overall, the SOCS box and SH2 domain show a conserved spatial domain arrangement with the BC box and substrate recognition domain of the von Hippel-Lindau (VHL) tumor suppressor protein, suggesting a common mechanism of ubiquitination in these cullin-dependent E3 ligases. The SOCS box binds elongin BC in a similar fashion to the VHL BC box and shows extended structural conservation with the F box of the Skp2 ubiquitin ligase. A previously unrecognized feature of the SOCS box is revealed with the burial of the C terminus, which packs together with the N-terminal extended SH2 subdomain to create a stable interface between the SOCS box and SH2 domain. This domain organization is conserved in SOCS1-3 and CIS1, which share a strictly conserved length of their C termini, but not in SOCS4, 5, and 7, which have extended C termini defining two distinct classes of inter- and intramolecular SOCS box interactions.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures



References
-
- Flores-Morales A., Greenhalgh C. J., Norstedt G., Rico-Bautista E. Mol. Endocrinol. 2005;20:241–253. - PubMed
-
- van Kerkhof P., Smeets M., Strous G. J. Endocrinology. 2002;143:1243–1252. - PubMed
-
- Zhang Y., Jiang J., Black R. A., Baumann G., Frank S. J. Endocrinology. 2000;141:4342–4348. - PubMed
-
- Ram P. A., Waxman D. J. J. Biol. Chem. 1997;272:17694–17702. - PubMed
-
- Metcalf D., Greenhalgh C. J., Viney E., Willson T. A., Starr R., Nicola N. A., Hilton D. J., Alexander W. S. Nature. 2000;405:1069–1073. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

