The bundling activity of vasodilator-stimulated phosphoprotein is required for filopodium formation
- PMID: 16675552
- PMCID: PMC1457090
- DOI: 10.1073/pnas.0511243103
The bundling activity of vasodilator-stimulated phosphoprotein is required for filopodium formation
Abstract
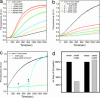
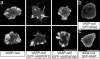
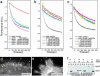
Filopodia are highly dynamic finger-like cell protrusions filled with parallel bundles of actin filaments. Previously we have shown that Diaphanous-related formin dDia2 is involved in the formation of filopodia. Another key player for the formation of filopodia across many species is vasodilator-stimulated phosphoprotein (VASP). It has been proposed that the essential role of VASP for formation of filopodia is its competition with capping proteins for filament barbed-end interaction. To better understand the function of VASP in filopodium formation, we analyzed the in vitro and in vivo properties of Dictyostelium VASP (DdVASP) and extended our findings to human VASP. Recombinant VASP from both species nucleated and bundled actin filaments, but did not compete with capping proteins or block depolymerization from barbed ends. Together with the finding that DdVASP binds to the FH2 domain of dDia2, these data indicate that the crucial role of VASP in filopodium formation is different from uncapping of actin filaments. To identify the activity of DdVASP required in this process, rescue experiments of DdVASP-null cells with mutant DdVASP constructs were performed. Only WT DdVASP, but not a mutant lacking the F-actin bundling activity, could rescue the ability of these cells to form WT-like filopodia. Our data suggest that DdVASP is complexed with dDia2 in filopodial tips and support formin-mediated filament elongation by bundling nascent actin filaments.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures




References
-
- Zigmond S. H. Curr. Opin. Cell Biol. 2004;16:99–105. - PubMed
-
- Watanabe N., Kato T., Fujita A., Ishizaki T., Narumiya S. Nat. Cell Biol. 1999;1:136–143. - PubMed
-
- Evangelista M., Pruyne D., Amberg D. C., Boone C., Bretscher A. Nat. Cell Biol. 2002;4:260–269. - PubMed
-
- Peng J., Wallar B. J., Flanders A., Swiatek P. J., Alberts A. S. Curr. Biol. 2003;13:534–545. - PubMed
-
- Pellegrin S., Mellor H. Curr. Biol. 2005;15:129–133. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

