Bayesian models for pooling microarray studies with multiple sources of replications
- PMID: 16677390
- PMCID: PMC1534062
- DOI: 10.1186/1471-2105-7-247
Bayesian models for pooling microarray studies with multiple sources of replications
Abstract
Background: Biologists often conduct multiple but different cDNA microarray studies that all target the same biological system or pathway. Within each study, replicate slides within repeated identical experiments are often produced. Pooling information across studies can help more accurately identify true target genes. Here, we introduce a method to integrate multiple independent studies efficiently.
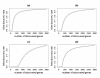
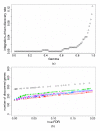
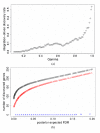
Results: We introduce a Bayesian hierarchical model to pool cDNA microarray data across multiple independent studies to identify highly expressed genes. Each study has multiple sources of variation, i.e. replicate slides within repeated identical experiments. Our model produces the gene-specific posterior probability of differential expression, which provides a direct method for ranking genes, and provides Bayesian estimates of false discovery rates (FDR). In simulations combining two and five independent studies, with fixed FDR levels, we observed large increases in the number of discovered genes in pooled versus individual analyses. When the number of output genes is fixed (e.g., top 100), the pooled model found appreciably more truly differentially expressed genes than the individual studies. We were also able to identify more differentially expressed genes from pooling two independent studies in Bacillus subtilis than from each individual data set. Finally, we observed that in our simulation studies our Bayesian FDR estimates tracked the true FDRs very well.
Conclusion: Our method provides a cohesive framework for combining multiple but not identical microarray studies with several sources of replication, with data produced from the same platform. We assume that each study contains only two conditions: an experimental and a control sample. We demonstrated our model's suitability for a small number of studies that have been either pre-scaled or have no outliers.
Figures

References
-
- Efron B, Tibshirani R, Storey JD, Tusher VG. Empirical Bayes Analysis of a Microarray Experiment. Journal of the American Statistical Association. 2001;96:1151–1160. doi: 10.1198/016214501753382129. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

