Core labeling of adenovirus with EGFP
- PMID: 16678874
- PMCID: PMC1781517
- DOI: 10.1016/j.virol.2006.03.042
Core labeling of adenovirus with EGFP
Abstract
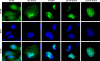
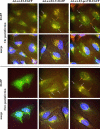
The study of adenovirus could greatly benefit from diverse methods of virus detection. Recently, it has been demonstrated that carboxy-terminal EGFP fusions of adenovirus core proteins Mu, V, and VII properly localize to the nucleus and display novel function in the cell. Based on these observations, we hypothesized that the core proteins may serve as targets for labeling the adenovirus core with fluorescent proteins. To this end, we constructed various chimeric expression vectors with fusion core genes (Mu-EGFP, V-EGFP, preVII-EGFP, and matVII-EGFP) while maintaining expression of the native proteins. Expression of the fusion core proteins was suboptimal using E1 expression vectors with both conventional CMV and modified (with adenovirus tripartite leader sequence) CMV5 promoters, resulting in non-labeled viral particles. However, robust expression equivalent to the native protein was observed when the fusion genes were placed in the deleted E3 region. The efficient Ad-wt-E3-V-EGFP and Ad-wt-E3-preVII-EGFP expression vectors were labeled allowing visualization of purified virus and tracking of the viral core during early infection. The vectors maintained their viral function, including viral DNA replication, viral DNA encapsidation, cytopathic effect, and thermostability. Core labeling offers a means to track the adenovirus core in vector targeting studies as well as basic adenovirus virology.
Figures






References
-
- Alemany R, Suzuki K, Curiel DT. Blood clearance rates of adenovirus type 5 in mice. J. Gen. Virol. 2000;81(Pt 11):2605–2609. - PubMed
-
- Alestrom P, Akusjarvi G, Lager M, Yeh-kai L, Pettersson U. Genes encoding the core proteins of adenovirus type 2. J. Biol. Chem. 1984;259(22):13980–13985. - PubMed
-
- Anderson CW, Young ME, Flint SJ. Characterization of the adenovirus 2 virion protein, mu. Virology. 1989;172(2):506–512. - PubMed
-
- Berk AJ. Adenovirus promoters and E1A transactivation. Annu. Rev. Genet. 1986;20:45–79. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

