pho2, a phosphate overaccumulator, is caused by a nonsense mutation in a microRNA399 target gene
- PMID: 16679417
- PMCID: PMC1489903
- DOI: 10.1104/pp.106.078063
pho2, a phosphate overaccumulator, is caused by a nonsense mutation in a microRNA399 target gene
Abstract
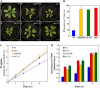
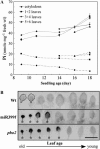
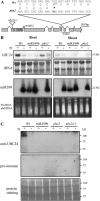
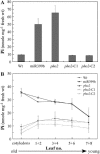
We recently demonstrated that microRNA399 (miR399) controls inorganic phosphate (Pi) homeostasis by regulating the expression of UBC24 encoding a ubiquitin-conjugating E2 enzyme in Arabidopsis (Arabidopsis thaliana). Transgenic plants overexpressing miR399 accumulated excessive Pi in the shoots and displayed Pi toxic symptoms. In this study, we revealed that a previously identified Pi overaccumulator, pho2, is caused by a single nucleotide mutation resulting in early termination within the UBC24 gene. The level of full-length UBC24 mRNA was reduced and no UBC24 protein was detected in the pho2 mutant, whereas up-regulation of miR399 by Pi deficiency was not affected. Several characteristics of Pi toxicity in the pho2 mutant were similar to those in the miR399-overexpressing and UBC24 T-DNA knockout plants: both Pi uptake and translocation of Pi from roots to shoots increased and Pi remobilization within leaves was impaired. These phenotypes of the pho2 mutation could be rescued by introduction of a wild-type copy of UBC24. Kinetic analyses revealed that greater Pi uptake in the pho2 and miR399-overexpressing plants is due to increased Vmax. The transcript level of most PHT1 Pi transporter genes was not significantly altered, except PHT1;8 whose expression was enhanced in Pi-sufficient roots of pho2 and miR399-overexpressing compared with wild-type plants. In addition, changes in the expression of several organelle-specific Pi transporters were noticed, which may be associated with the redistribution of intracellular Pi under excess Pi. Furthermore, miR399 and UBC24 were colocalized in the vascular cylinder. This observation not only provides important insight into the interaction between miR399 and UBC24 mRNA, but also supports their systemic function in Pi translocation and remobilization.
Figures


Comment in
-
Phosphate accumulation in plants: signaling.Plant Physiol. 2008 Sep;148(1):3-5. doi: 10.1104/pp.104.900269. Plant Physiol. 2008. PMID: 18772350 Free PMC article. No abstract available.
References
-
- Allen E, Xie Z, Gustafson AM, Carrington JC (2005) microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121: 207–221 - PubMed
-
- Ames BN (1966) Assay of inorganic phosphate, total phosphate and phosphatases. Methods Enzymol 8: 115–118
-
- Barber SA, Walker JM, Vasey EH (1963) Mechanisms for the movement of plant nutrients from the soil and fertilizer to the plant root. J Agric Food Chem 11: 204–207
-
- Bieleski RL (1973) Phosphate pools, phosphate transport, and phosphate availability. Annu Rev Plant Physiol 24: 225–252
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

