PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants
- PMID: 16679424
- PMCID: PMC1489890
- DOI: 10.1104/pp.106.079707
PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants
Abstract
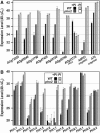
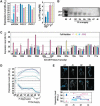
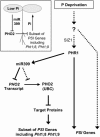
Inorganic phosphate (Pi)-signaling pathways in plants are still largely unknown. The Arabidopsis (Arabidopsis thaliana) pho2 mutant overaccumulates Pi in leaves in Pi-replete conditions. Micrografting revealed that a pho2 root genotype is sufficient to yield leaf Pi accumulation. In pho2 mutants, Pi does not repress a set of Pi starvation-induced genes, including AtIPS1, AT4, and Pi transporters Pht1;8 and Pht1;9. Map-based cloning identified PHO2 as At2g33770, an unusual E2 conjugase gene. It was recently shown that Pi deprivation induces mature microRNA (miRNA [miR399]) and that overexpression of miR399 in Pi-replete conditions represses E2 conjugase expression and leads to high leaf Pi concentrations, thus phenocopying pho2. We show here that miR399 primary transcripts are also strongly induced by low Pi and rapidly repressed after addition of Pi. PHO2 transcripts change reciprocally to miR399 transcripts in Pi-deprived plants and in miR399 overexpressers. However, responses after Pi readdition and in beta-glucuronidase reporter lines suggest that PHO2 expression is also regulated by Pi in a manner unrelated to miR399-mediated transcript cleavage. Expression of miR399 was strongly reduced in Pi-deprived Arabidopsis phr1 mutants, and a subset of Pi-responsive genes repressed in Pi-deprived phr1 mutants was up-regulated in Pi-replete pho2 mutants. This places miR399 and PHO2 in a branch of the Pi-signaling network downstream of PHR1. Finally, putative PHO2 orthologs containing five miR399-binding sites in their 5'-untranslated regions were identified in other higher plants, and Pi-dependent miR399 expression was demonstrated in rice (Oryza sativa), suggesting a conserved regulatory mechanism.
Figures

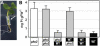


Comment in
-
Phosphate accumulation in plants: signaling.Plant Physiol. 2008 Sep;148(1):3-5. doi: 10.1104/pp.104.900269. Plant Physiol. 2008. PMID: 18772350 Free PMC article. No abstract available.
References
-
- Abel S, Ticconi CA, Delatorre CA (2002) Phosphate sensing in higher plants. Physiol Plant 115: 1–8 - PubMed
-
- Allen E, Xie Z, Gustafson AM, Carrington JC (2005) microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121: 207–221 - PubMed
-
- Bolstad BM, Irizarry RA, Astrand M, Speed TP (2003) A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19: 185–193 - PubMed
-
- Chen DL, Delatorre CA, Bakker A, Abel S (2000) Conditional identification of phosphate-starvation-response mutants in Arabidopsis thaliana. Planta 211: 13–22 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

