High-resolution recombination patterns in a region of human chromosome 21 measured by sperm typing
- PMID: 16680198
- PMCID: PMC1456319
- DOI: 10.1371/journal.pgen.0020070
High-resolution recombination patterns in a region of human chromosome 21 measured by sperm typing
Abstract
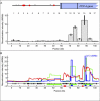
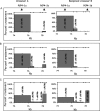
For decades, classical crossover studies and linkage disequilibrium (LD) analysis of genomic regions suggested that human meiotic crossovers may not be randomly distributed along chromosomes but are focused instead in "hot spots." Recent sperm typing studies provided data at very high resolution and accuracy that defined the physical limits of a number of hot spots. The data were also used to test whether patterns of LD can predict hot spot locations. These sperm typing studies focused on several small regions of the genome already known or suspected of containing a hot spot based on the presence of LD breakdown or previous experimental evidence of hot spot activity. Comparable data on target regions not specifically chosen using these two criteria is lacking but is needed to make an unbiased test of whether LD data alone can accurately predict active hot spots. We used sperm typing to estimate recombination in 17 almost contiguous ~5 kb intervals spanning 103 kb of human Chromosome 21. We found two intervals that contained new hot spots. The comparison of our data with recombination rates predicted by statistical analyses of LD showed that, overall, the two datasets corresponded well, except for one predicted hot spot that showed little crossing over. This study doubles the experimental data on recombination in men at the highest resolution and accuracy and supports the emerging genome-wide picture that recombination is localized in small regions separated by cold areas. Detailed study of one of the new hot spots revealed a sperm donor with a decrease in recombination intensity at the canonical recombination site but an increase in crossover activity nearby. This unique finding suggests that the position and intensity of hot spots may evolve by means of a concerted mechanism that maintains the overall recombination intensity in the region.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures


References
-
- Kauppi L, Jeffreys AJ, Keeney S. Where the crossovers are: Recombination distributions in mammals. Nat Rev Genet. 2004;5:413–424. - PubMed
-
- Petes TD. Meiotic recombination hot spots and cold spots. Nat Rev Genet. 2001;2:360–369. - PubMed
-
- Carrington M, Cullen M. Justified chauvinism: Advances in defining meiotic recombination through sperm typing. Trends Genet. 2004;20:196–205. - PubMed
-
- Clark AG. Hot spots unglued. Nat Genet. 2005;37:563–564. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

