Preferential distribution of active RNA polymerase II molecules in the nuclear periphery
- PMID: 1668144
- PMCID: PMC5952200
Preferential distribution of active RNA polymerase II molecules in the nuclear periphery
Abstract
We have combined immunogold labeling with the Miller spreading technique in order to localize proteins at the electron microscope (EM) level in whole mount nuclei from mouse and human fibroblasts. Anti-histone H1 antibody labels nuclei uniformly, indicating that the nuclear interior is accessible to both antibodies and gold conjugates. Anti-topoisomerase I antibody labels nucleoli intensely, in agreement with previous immunofluorescent and biochemical data. Two different antibodies against the large subunit of RNA polymerase II (pol II) show preferential labeling of the nuclear periphery, as do antibodies against lamin, a known peripheral nuclear protein. Treatment of cells with alpha-amanitin results in loss of virtually all RNA polymerase II staining, supporting the specificity of labeling. Finally, when nuclei are incubated in the presence of biotin-UTP (bio-UTP) under run-off transcription conditions, incorporation is preferentially located at the nuclear periphery. These results support the conclusions that transcriptionally active pol II molecules are non-uniformly distributed in fibroblast nuclei, and that their differential distribution mirrors that of total pol II.
Figures
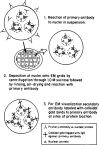
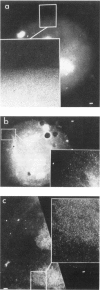
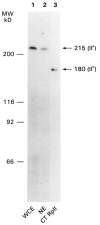

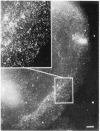
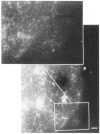
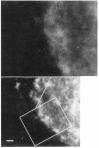
Similar articles
-
Lamin A/C speckles mediate spatial organization of splicing factor compartments and RNA polymerase II transcription.J Cell Biol. 2002 Dec 9;159(5):783-93. doi: 10.1083/jcb.200204149. Epub 2002 Dec 9. J Cell Biol. 2002. PMID: 12473687 Free PMC article.
-
Discrete localization of different DNA topoisomerases in HeLa and K562 cell nuclei and subnuclear fractions.Exp Cell Res. 1994 Feb;210(2):336-48. doi: 10.1006/excr.1994.1046. Exp Cell Res. 1994. PMID: 8299728
-
Nuclear distribution of Oct-4 transcription factor in transcriptionally active and inactive mouse oocytes and its relation to RNA polymerase II and splicing factors.J Cell Biochem. 2003 Jul 1;89(4):720-32. doi: 10.1002/jcb.10545. J Cell Biochem. 2003. PMID: 12858338
-
Transcription-dependent redistribution of the large subunit of RNA polymerase II to discrete nuclear domains.J Cell Biol. 1995 Apr;129(2):287-98. doi: 10.1083/jcb.129.2.287. J Cell Biol. 1995. PMID: 7536746 Free PMC article.
-
Mapping proteins to nuclear pore complexes by immunogold electron microscopy.Methods Cell Biol. 1998;53:287-302. doi: 10.1016/s0091-679x(08)60883-3. Methods Cell Biol. 1998. PMID: 9348513 Review. No abstract available.
Cited by
-
Analysis of spatial point patterns in nuclear biology.PLoS One. 2012;7(5):e36841. doi: 10.1371/journal.pone.0036841. Epub 2012 May 16. PLoS One. 2012. PMID: 22615822 Free PMC article.
-
Preferential repair of cyclobutane pyrimidine dimers in the transcribed strand of a gene in yeast chromosomes and plasmids is dependent on transcription.Proc Natl Acad Sci U S A. 1992 Nov 15;89(22):10696-700. doi: 10.1073/pnas.89.22.10696. Proc Natl Acad Sci U S A. 1992. PMID: 1438266 Free PMC article.
-
In vivo degradation of RNA polymerase II largest subunit triggered by alpha-amanitin.Nucleic Acids Res. 1996 Aug 1;24(15):2924-9. doi: 10.1093/nar/24.15.2924. Nucleic Acids Res. 1996. PMID: 8760875 Free PMC article.
-
Fractionation of human H1 subtypes and characterization of a subtype-specific antibody exhibiting non-uniform nuclear staining.Chromosome Res. 1993 Jul;1(2):127-39. doi: 10.1007/BF00710036. Chromosome Res. 1993. PMID: 7511470
-
Reverse recruitment: the Nup84 nuclear pore subcomplex mediates Rap1/Gcr1/Gcr2 transcriptional activation.Proc Natl Acad Sci U S A. 2005 Apr 19;102(16):5749-54. doi: 10.1073/pnas.0501768102. Epub 2005 Apr 7. Proc Natl Acad Sci U S A. 2005. PMID: 15817685 Free PMC article.
References
-
- Agard D. and Sedat J. (1983), Nature 302, 676–681. - PubMed
-
- Cho K. W. Y., Khalili K., Zanndmeni R., and Weinmann R. (1985), J Biol Chem 260, 15204–15210. - PubMed
-
- Cockerill P. N. and Garrard W. T. (1986), Cell 44, 273–282. - PubMed
-
- Cremer T., Cremer C., Baumann H., Leudtke E.-K., Sperling K., Teuber V., and Zorn C. (1982) Hum Genet 60, 40–56. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
