Use of miniaturized protein arrays for Escherichia coli O serotyping
- PMID: 16682477
- PMCID: PMC1459650
- DOI: 10.1128/CVI.13.5.561-567.2006
Use of miniaturized protein arrays for Escherichia coli O serotyping
Abstract
Serological typing of Escherichia coli O antigens is a well-established method used for differentiation and identification of O serotypes commonly associated with disease. In this feasibility study, we have developed a novel somatic antibody-based miniaturized microarray chip, using 17 antisera, which can be used to detect bound whole-cell E. coli antigen with its corresponding immobilized antibody, to assess the feasibility of this approach. The chip was tested using the related 17 control strains, and the O types found by the microarray chip showed 100% correlation with the O types found by conventional typing. A blind trial was performed in which 100 E. coli isolates that had been O serotyped previously by the conventional assay were tested by the array approach. Overall, the O serotypes of 88% of isolates were correctly identified by the microarray method. For several isolates, ambiguity of O-type designation by microarray arose due to increased sensitivity of this method, allowing signal intensities of cross-reactions to be quantified. Investigation of discrepancies between conventional and microarray O serotyping indicated that some isolates upon storage had become untypeable and, therefore, gave poor signal intensity when tested by the microarray or retested by conventional means. For all 20 serotype O26 and O157 isolates, the apparent discrepancy in O serotyping was analyzed further by a third independent test, which confirmed the microarray results. Therefore, the use of miniaturized protein arrays increases the speed and efficiency of O serotyping in a cost-effective manner, and these preliminary findings suggest the microarray approach may have a higher accuracy than those of traditional O-serotyping methods.
Figures


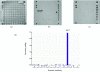
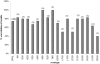
References
-
- Aktan, I., K. A. Sprigings, R. M. La Ragione, L. M. Faulkner, G. A. Paiba, and M. J. Woodward. 2004. Characterisation of attaching-effacing Escherichia coli isolated from animals at slaughter in England and Wales. Vet. Microbiol. 102:43-53. - PubMed
-
- Di Padova, F. E., H. Brade, G. R. Barclay, I. R. Poxton, E. Liehl, E. Schuetze, H. P. Kocher, G. Ramsay, M. H. Schreier, D. B. McClelland, and E. T. Rietschel. 1993. A broadly cross-protective monoclonal antibody binding to Escherichia coli and Salmonella lipopolysaccharides. Infect. Immun. 61:3863-3872. - PMC - PubMed
-
- Ewing, W. H. 1986. The genus Escherichia, p. 93-136. In W. H. Ewing (ed.), Edwards and Ewings identification of Enterobacteriaceae, 4th ed. Elsevier Science Publishing Co. Inc., New York, N.Y.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

