Maintenance of viral suppression in HIV-1-infected HLA-B*57+ elite suppressors despite CTL escape mutations
- PMID: 16682496
- PMCID: PMC2121215
- DOI: 10.1084/jem.20052319
Maintenance of viral suppression in HIV-1-infected HLA-B*57+ elite suppressors despite CTL escape mutations
Abstract
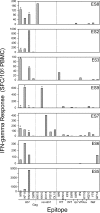
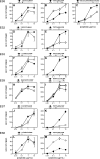
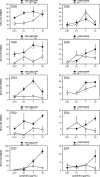
Rare human immunodeficiency virus 1-infected individuals, termed elite suppressors (ES), maintain plasma virus levels of <50 copies/ml and normal CD4 counts without therapy. The major histocompatibility complex class I allele group human histocompatibility leukocyte antigen (HLA)-B*57 is overrepresented in this population. Mutations in HLA-B*57-restricted epitopes have been observed in ES, but their significance has remained unclear. Here we investigate the extent and impact of cytotoxic T lymphocyte (CTL) escape mutations in HLA-B*57+ ES. We provide the first direct evidence that most ES experience chronic low level viremia. Sequencing revealed a striking discordance between the genotypes of plasma virus and archived provirus in resting CD4+ T cells. Mutations in HLA-B*57-restricted Gag epitopes were present in all viruses from plasma but were rare in proviruses, suggesting powerful selective pressure acting at these epitopes. Surprisingly, strong CD8+ T cell interferon-gamma responses were detected against some mutant epitopes found in plasma virus, suggesting the development of de novo responses to viral variants. In some individuals, relative CD8+ T cell interleukin-2 responses showed better correlation with the selection observed in vivo. Thus, analysis of low level viremia reveals an unexpectedly high level of CTL escape mutations reflecting selective pressure acting at HLA-B*57-restricted epitopes in ES. Continued viral suppression probably reflects CTL responses against unmutated epitopes and residual or de novo responses against epitopes with escape mutations.
Figures



References
-
- Schmitz, J.E., M.J. Kuroda, S. Santra, V.G. Sasseville, M.A. Simon, M.A. Lifton, P. Racz, K. Tenner-Racz, M. Dalesandro, B.J. Scallon, et al. 1999. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science. 283:857–860. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

