Codon usage domains over bacterial chromosomes
- PMID: 16683018
- PMCID: PMC1447655
- DOI: 10.1371/journal.pcbi.0020037
Codon usage domains over bacterial chromosomes
Abstract
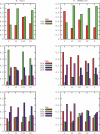
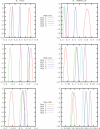
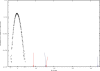
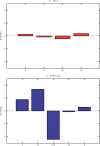
The geography of codon bias distributions over prokaryotic genomes and its impact upon chromosomal organization are analyzed. To this aim, we introduce a clustering method based on information theory, specifically designed to cluster genes according to their codon usage and apply it to the coding sequences of Escherichia coli and Bacillus subtilis. One of the clusters identified in each of the organisms is found to be related to expression levels, as expected, but other groups feature an over-representation of genes belonging to different functional groups, namely horizontally transferred genes, motility, and intermediary metabolism. Furthermore, we show that genes with a similar bias tend to be close to each other on the chromosome and organized in coherent domains, more extended than operons, demonstrating a role of translation in structuring bacterial chromosomes. It is argued that a sizeable contribution to this effect comes from the dynamical compartimentalization induced by the recycling of tRNAs, leading to gene expression rates dependent on their genomic and expression context.
Conflict of interest statement
Figures


References
-
- Post L, Nomura M. DNA sequences from the str operon of Escherichia coli. J Biol Chem 255: 4660–4666; 1980. Available: http://www.jbc.org/cgi/content/abstract/255/10/4660. Accessed 20 March 2006. - PubMed
-
- Ikemura T. Correlation between the abundance of Escherichia coli tRNAs and the occurrence of the respective codons in its protein genes. J Mol Biol. 1981;146:1–21. - PubMed
-
- Ikemura T. Correlation between the abundance of yeast tRNAs and the occurrence of the respective codons in protein genes: Differences in synonymous codon choice patterns of yeast and Escherichia coli with reference to the abundance of isoaccepting tRNAs. J Mol Biol. 1982;158:573–597. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

