Heterotachy in mammalian promoter evolution
- PMID: 16683025
- PMCID: PMC1449885
- DOI: 10.1371/journal.pgen.0020030
Heterotachy in mammalian promoter evolution
Abstract
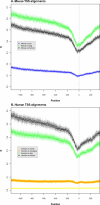
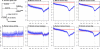
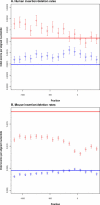
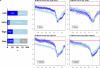
We have surveyed the evolutionary trends of mammalian promoters and upstream sequences, utilising large sets of experimentally supported transcription start sites (TSSs). With 30,969 well-defined TSSs from mouse and 26,341 from human, there are sufficient numbers to draw statistically meaningful conclusions and to consider differences between promoter types. Unlike previous smaller studies, we have considered the effects of insertions, deletions, and transposable elements as well as nucleotide substitutions. The rate of promoter evolution relative to that of control sequences has not been consistent between lineages nor within lineages over time. The most pronounced manifestation of this heterotachy is the increased rate of evolution in primate promoters. This increase is seen across different classes of mutation, including substitutions and micro-indel events. We investigated the relationship between promoter and coding sequence selective constraint and suggest that they are generally uncorrelated. This analysis also identified a small number of mouse promoters associated with the immune response that are under positive selection in rodents. We demonstrate significant differences in divergence between functional promoter categories and identify a category of promoters, not associated with conventional protein-coding genes, that has the highest rates of divergence across mammals. We find that evolutionary rates vary both on a fine scale within mammalian promoters and also between different functional classes of promoters. The discovery of heterotachy in promoter evolution, in particular the accelerated evolution of primate promoters, has important implications for our understanding of human evolution and for strategies to detect primate-specific regulatory elements.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

