Relaxed phylogenetics and dating with confidence
- PMID: 16683862
- PMCID: PMC1395354
- DOI: 10.1371/journal.pbio.0040088
Relaxed phylogenetics and dating with confidence
Abstract
In phylogenetics, the unrooted model of phylogeny and the strict molecular clock model are two extremes of a continuum. Despite their dominance in phylogenetic inference, it is evident that both are biologically unrealistic and that the real evolutionary process lies between these two extremes. Fortunately, intermediate models employing relaxed molecular clocks have been described. These models open the gate to a new field of "relaxed phylogenetics." Here we introduce a new approach to performing relaxed phylogenetic analysis. We describe how it can be used to estimate phylogenies and divergence times in the face of uncertainty in evolutionary rates and calibration times. Our approach also provides a means for measuring the clocklikeness of datasets and comparing this measure between different genes and phylogenies. We find no significant rate autocorrelation among branches in three large datasets, suggesting that autocorrelated models are not necessarily suitable for these data. In addition, we place these datasets on the continuum of clocklikeness between a strict molecular clock and the alternative unrooted extreme. Finally, we present analyses of 102 bacterial, 106 yeast, 61 plant, 99 metazoan, and 500 primate alignments. From these we conclude that our method is phylogenetically more accurate and precise than the traditional unrooted model while adding the ability to infer a timescale to evolution.
Figures
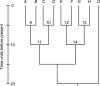
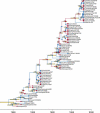
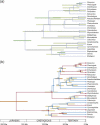
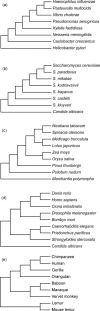
Comment in
-
Relaxing the clock brings time back into phylogenetics.PLoS Biol. 2006 May;4(5):e106. doi: 10.1371/journal.pbio.0040106. Epub 2006 Mar 14. PLoS Biol. 2006. PMID: 20076564 Free PMC article. No abstract available.
-
The dawn of relaxed phylogenetics.PLoS Biol. 2023 Jan 25;21(1):e3001998. doi: 10.1371/journal.pbio.3001998. eCollection 2023 Jan. PLoS Biol. 2023. PMID: 36696649 Free PMC article.
References
-
- Zuckerkandl E, Pauling L. Molecular disease, evolution and genic heterogeneity. In: Kasha M, Pullman B, editors. Horizons in biochemistry. New York: Academic Press; 1962. pp. 189–225.
-
- Zuckerkandl E, Pauling L. Evolutionary divergence and convergence in proteins. In: Bryson V, Vogel HJ, editors. Evolving genes and proteins. New York: Academic Press; 1965. pp. 97–166.
-
- Britten RJ. Rates of DNA sequence evolution differ between taxonomic groups. Science. 1986;231:1393–1398. - PubMed
-
- Hasegawa M, Kishino H. Heterogeneity of tempo and mode of mitochondrial DNA evolution among mammalian orders. Jpn J Genet. 1989;64:243–258. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

