Calcium-dependent and -independent interactions of the S100 protein family
- PMID: 16683912
- PMCID: PMC1462724
- DOI: 10.1042/BJ20060195
Calcium-dependent and -independent interactions of the S100 protein family
Abstract
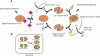
The S100 proteins comprise at least 25 members, forming the largest group of EF-hand signalling proteins in humans. Although the proteins are expressed in many tissues, each S100 protein has generally been shown to have a preference for expression in one particular tissue or cell type. Three-dimensional structures of several S100 family members have shown that the proteins assume a dimeric structure consisting of two EF-hand motifs per monomer. Calcium binding to these S100 proteins, with the exception of S100A10, results in an approx. 40 degrees alteration in the position of helix III, exposing a broad hydrophobic surface that enables the S100 proteins to interact with a variety of target proteins. More than 90 potential target proteins have been documented for the S100 proteins, including the cytoskeletal proteins tubulin, glial fibrillary acidic protein and F-actin, which have been identified mostly from in vitro experiments. In the last 5 years, efforts have concentrated on quantifying the protein interactions of the S100 proteins, identifying in vivo protein partners and understanding the molecular specificity for target protein interactions. Furthermore, the S100 proteins are the only EF-hand proteins that are known to form both homo- and hetero-dimers, and efforts are underway to determine the stabilities of these complexes and structural rationales for their formation and potential differences in their biological roles. This review highlights both the calcium-dependent and -independent interactions of the S100 proteins, with a focus on the structures of the complexes, differences and similarities in the strengths of the interactions, and preferences for homo- compared with hetero-dimeric S100 protein assembly.
Figures
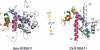

References
-
- Berridge M. J., Lipp P., Bootman M. D. The versatility and universality of calcium signalling. Nat. Rev. Mol. Cell Biol. 2000;1:11–21. - PubMed
-
- Berridge M. J., Bootman M. D., Roderick H. L. Calcium signalling: dynamics, homeostasis and remodelling. Nat. Rev. Mol. Cell Biol. 2003;4:517–529. - PubMed
-
- Kretsinger R. H., Nockolds C. E. Carp muscle calcium-binding protein. II. Structure determination and general description. J. Biol. Chem. 1973;248:3313–3326. - PubMed
-
- Strynadka N. C. J., James M. N. G. Crystal structures of the helix–loop–helix calcium-binding proteins. Annu. Rev. Biochem. 1989;58:951–998. - PubMed
-
- Marsden B. J., Shaw G. S., Sykes B. D. Calcium binding proteins: elucidating the contributions to calcium affinity from analysis of species variants and peptide fragments. Biochem. Cell Biol. 1990;68:587–601. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

