RNA interference is not involved in natural antisense mediated regulation of gene expression in mammals
- PMID: 16684369
- PMCID: PMC1779516
- DOI: 10.1186/gb-2006-7-5-r38
RNA interference is not involved in natural antisense mediated regulation of gene expression in mammals
Abstract
Background: Antisense transcription, yielding both coding and non-coding RNA, is a widespread phenomenon in mammals. The mechanism by which natural antisense transcripts (NAT) may regulate gene expression are largely unknown. The aim of the present study was to explore the mechanism of reciprocal sense-antisense (S-AS) regulation by studying the effects of a coding and non-coding NAT on corresponding gene expression, and to investigate the possible involvement of endogenous RNA interference (RNAi) in S-AS interactions.
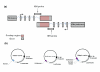
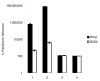
Results: We have examined the mechanism of S-AS RNA base pairing, using thymidylate synthase and hypoxia inducible factor-1alpha as primary examples of endogenous genes with coding and non-coding NAT partners, respectively. Here we provide direct evidence against S-AS RNA duplex formation in the cytoplasm of human cells and subsequent activation of RNAi.
Conclusion: Collectively, our data demonstrate that NAT regulation of gene expression occurs through a pathway independent of Dicer associated RNAi. Moreover, we introduce an experimental strategy with utility for the functional examination of other S-AS pair interactions.
Figures





References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

