Influence of metabolic network structure and function on enzyme evolution
- PMID: 16684370
- PMCID: PMC1779518
- DOI: 10.1186/gb-2006-7-5-r39
Influence of metabolic network structure and function on enzyme evolution
Abstract
Background: Most studies of molecular evolution are focused on individual genes and proteins. However, understanding the design principles and evolutionary properties of molecular networks requires a system-wide perspective. In the present work we connect molecular evolution on the gene level with system properties of a cellular metabolic network. In contrast to protein interaction networks, where several previous studies investigated the molecular evolution of proteins, metabolic networks have a relatively well-defined global function. The ability to consider fluxes in a metabolic network allows us to relate the functional role of each enzyme in a network to its rate of evolution.
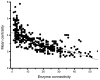
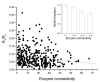
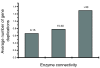
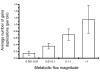
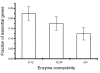
Results: Our results, based on the yeast metabolic network, demonstrate that important evolutionary processes, such as the fixation of single nucleotide mutations, gene duplications, and gene deletions, are influenced by the structure and function of the network. Specifically, central and highly connected enzymes evolve more slowly than less connected enzymes. Also, enzymes carrying high metabolic fluxes under natural biological conditions experience higher evolutionary constraints. Genes encoding enzymes with high connectivity and high metabolic flux have higher chances to retain duplicates in evolution. In contrast to protein interaction networks, highly connected enzymes are no more likely to be essential compared to less connected enzymes.
Conclusion: The presented analysis of evolutionary constraints, gene duplication, and essentiality demonstrates that the structure and function of a metabolic network shapes the evolution of its enzymes. Our results underscore the need for systems-based approaches in studies of molecular evolution.
Figures
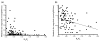
Comment in
-
Impacts of yeast metabolic network structure on enzyme evolution.Genome Biol. 2007;8(8):407. doi: 10.1186/gb-2007-8-8-407. Genome Biol. 2007. PMID: 17692134 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

