Multilocus association mapping using variable-length Markov chains
- PMID: 16685642
- PMCID: PMC1474089
- DOI: 10.1086/503876
Multilocus association mapping using variable-length Markov chains
Abstract
I propose a new method for association-based gene mapping that makes powerful use of multilocus data, is computationally efficient, and is straightforward to apply over large genomic regions. The approach is based on the fitting of variable-length Markov chain models, which automatically adapt to the degree of linkage disequilibrium (LD) between markers to create a parsimonious model for the LD structure. Edges of the fitted graph are tested for association with trait status. This approach can be thought of as haplotype testing with sophisticated windowing that accounts for extent of LD to reduce degrees of freedom and number of tests while maximizing information. I present analyses of two published data sets that show that this approach can have better power than single-marker tests or sliding-window haplotypic tests.
Figures

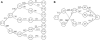
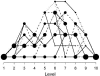

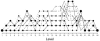

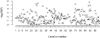
References
Web Resources
-
- HapVLMC, http://www.stat.auckland.ac.nz/~browning/HapVLMC/index.htm (for R code for implementing the proposed method)
-
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for cystic fibrosis and Crohn disease)
References
-
- Excoffier L, Slatkin M (1995) Maximum-likelihood estimation of molecular haplotype frequencies in a diploid population. Mol Biol Evol 12:921–927 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

