Reconstruction of a functional human gene network, with an application for prioritizing positional candidate genes
- PMID: 16685651
- PMCID: PMC1474084
- DOI: 10.1086/504300
Reconstruction of a functional human gene network, with an application for prioritizing positional candidate genes
Abstract
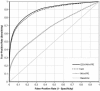
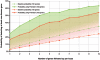
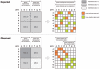
Most common genetic disorders have a complex inheritance and may result from variants in many genes, each contributing only weak effects to the disease. Pinpointing these disease genes within the myriad of susceptibility loci identified in linkage studies is difficult because these loci may contain hundreds of genes. However, in any disorder, most of the disease genes will be involved in only a few different molecular pathways. If we know something about the relationships between the genes, we can assess whether some genes (which may reside in different loci) functionally interact with each other, indicating a joint basis for the disease etiology. There are various repositories of information on pathway relationships. To consolidate this information, we developed a functional human gene network that integrates information on genes and the functional relationships between genes, based on data from the Kyoto Encyclopedia of Genes and Genomes, the Biomolecular Interaction Network Database, Reactome, the Human Protein Reference Database, the Gene Ontology database, predicted protein-protein interactions, human yeast two-hybrid interactions, and microarray co-expressions. We applied this network to interrelate positional candidate genes from different disease loci and then tested 96 heritable disorders for which the Online Mendelian Inheritance in Man database reported at least three disease genes. Artificial susceptibility loci, each containing 100 genes, were constructed around each disease gene, and we used the network to rank these genes on the basis of their functional interactions. By following up the top five genes per artificial locus, we were able to detect at least one known disease gene in 54% of the loci studied, representing a 2.8-fold increase over random selection. This suggests that our method can significantly reduce the cost and effort of pinpointing true disease genes in analyses of disorders for which numerous loci have been reported but for which most of the genes are unknown.
Figures
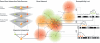



References
Web Resources
-
- Biomolecular Interaction Network Database (BIND), http://bind.ca/
-
- Ensembl, http://www.ensembl.org/index.html
-
- GeneNetwork, http://www.genenetwork.nl
-
- Human Protein Reference Database (HPRD), http://www.hprd.org/
-
- Kyoto Encyclopedia of Genes and Genomes (KEGG), http://www.genome.jp/kegg/
References
-
- Seri M, Martucciello G, Paleari L, Bolino A, Priolo M, Salemi G, Forabosco P, Caroli F, Cusano R, Tocco T, Lerone M, Cama A, Torre M, Guys JM, Romeo G, Jasonni V (1999) Exclusion of the Sonic Hedgehog gene as responsible for Currarino syndrome and anorectal malformations with sacral hypodevelopment. Hum Genet 104:108–11010.1007/s004390050919 - DOI - PubMed
-
- Simard J, Feunteun J, Lenoir G, Tonin P, Normand T, Luu The V, Vivier A, et al (1993) Genetic mapping of the breast-ovarian cancer syndrome to a small interval on chromosome 17q12-21: exclusion of candidate genes EDH17B2 and RARA. Hum Mol Genet 2:1193–1199 - PubMed
-
- Tumer Z, Croucher PJ, Jensen LR, Hampe J, Hansen C, Kalscheuer V, Ropers HH, Tommerup N, Schreiber S (2002) Genomic structure, chromosome mapping and expression analysis of the human AVIL gene, and its exclusion as a candidate for locus for inflammatory bowel disease at 12q13-14 (IBD2). Gene 288:179–18510.1016/S0378-1119(02)00478-X - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

