The Inferelator: an algorithm for learning parsimonious regulatory networks from systems-biology data sets de novo
- PMID: 16686963
- PMCID: PMC1779511
- DOI: 10.1186/gb-2006-7-5-r36
The Inferelator: an algorithm for learning parsimonious regulatory networks from systems-biology data sets de novo
Abstract
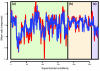
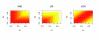
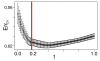
We present a method (the Inferelator) for deriving genome-wide transcriptional regulatory interactions, and apply the method to predict a large portion of the regulatory network of the archaeon Halobacterium NRC-1. The Inferelator uses regression and variable selection to identify transcriptional influences on genes based on the integration of genome annotation and expression data. The learned network successfully predicted Halobacterium's global expression under novel perturbations with predictive power similar to that seen over training data. Several specific regulatory predictions were experimentally tested and verified.
Figures

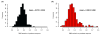
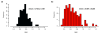
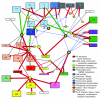
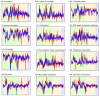
References
-
- Herrgard MJ, Covert MW, Palsson BO. Reconstruction of microbial transcriptional regulatory networks. Curr Opin Biotechnol. 2004;15:70–77. - PubMed
-
- De Jong H. Modeling and simulation of genetic regulatory systems: a literature review. J Comput Biol. 2002;9:67–103. - PubMed
-
- Alm E, Arkin AP. Biological networks. Curr Opin Struct Biol. 2003;13:193–202. - PubMed
-
- Hashimoto RF, Kim S, Shmulevich I, Zhang W, Bittner ML, Dougherty ER. Growing genetic regulatory networks from seed genes. Bioinformatics. 2004;20:1241–1247. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

