The two eIF4A helicases in Trypanosoma brucei are functionally distinct
- PMID: 16687655
- PMCID: PMC1459412
- DOI: 10.1093/nar/gkl290
The two eIF4A helicases in Trypanosoma brucei are functionally distinct
Abstract
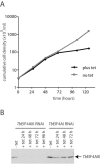
Protozoan parasites belonging to the family Trypanosomatidae are characterized by an unusual pathway for the production of mRNAs via polycistronic transcription and trans-splicing of a 5' capped mini-exon which is linked to the 3' cleavage and polyadenylation of the upstream transcript. However, little is known of the mechanism of protein synthesis in these organisms, despite their importance as agents of a number of human diseases. Here we have investigated the role of two Trypanosoma brucei homologues of the translation initiation factor eIF4A (in the light of subsequent experiments these were named as TbEIF4AI and TbEIF4AIII). eIF4A, a DEAD-box RNA helicase, is a subunit of the translation initiation complex eIF4F which binds to the cap structure of eukaryotic mRNA and recruits the small ribosomal subunit. TbEIF4AI is a very abundant predominantly cytoplasmic protein (over 1 x 10(5) molecules/cell) and depletion to approximately 10% of normal levels through RNA interference dramatically reduces protein synthesis one cell cycle following double-stranded RNA induction and stops cell proliferation. In contrast, TbEIF4AIII is a nuclear, moderately expressed protein (approximately 1-2 x 10(4) molecules/cell), and its depletion stops cellular proliferation after approximately four cell cycles. Ectopic expression of a dominant negative mutant of TbEIF4AI, but not of TbEIF4AIII, induced a slow growth phenotype in transfected cells. Overall, our results suggest that only TbEIF4AI is involved in protein synthesis while the properties and sequence of TbEIF4AIII indicate that it may be the orthologue of eIF4AIII, a component of the exon junction complex in mammalian cells.
Figures







Similar articles
-
Distinct modes of interaction within eIF4F-like complexes and susceptibility to the RocA inhibitor for the Trypanosoma brucei EIF4AI translation initiation factor.PLoS One. 2025 May 9;20(5):e0322812. doi: 10.1371/journal.pone.0322812. eCollection 2025. PLoS One. 2025. PMID: 40343969 Free PMC article.
-
Two related trypanosomatid eIF4G homologues have functional differences compatible with distinct roles during translation initiation.RNA Biol. 2015;12(3):305-19. doi: 10.1080/15476286.2015.1017233. RNA Biol. 2015. PMID: 25826663 Free PMC article.
-
tbCPSF30 depletion by RNA interference disrupts polycistronic RNA processing in Trypanosoma brucei.J Biol Chem. 2003 Jul 18;278(29):26870-8. doi: 10.1074/jbc.M302405200. Epub 2003 May 13. J Biol Chem. 2003. PMID: 12746436
-
Eukaryotic initiation factor 4A (eIF4A) during viral infections.Virus Genes. 2019 Jun;55(3):267-273. doi: 10.1007/s11262-019-01641-7. Epub 2019 Feb 22. Virus Genes. 2019. PMID: 30796742 Free PMC article. Review.
-
The diverse roles of the eIF4A family: you are the company you keep.Biochem Soc Trans. 2014 Feb;42(1):166-72. doi: 10.1042/BST20130161. Biochem Soc Trans. 2014. PMID: 24450646 Review.
Cited by
-
Invariant surface glycoprotein 65 of Trypanosoma brucei is a complement C3 receptor.Nat Commun. 2022 Aug 29;13(1):5085. doi: 10.1038/s41467-022-32728-9. Nat Commun. 2022. PMID: 36038546 Free PMC article.
-
The DEAD-box helicase eIF4A: paradigm or the odd one out?RNA Biol. 2013 Jan;10(1):19-32. doi: 10.4161/rna.21966. Epub 2012 Sep 20. RNA Biol. 2013. PMID: 22995829 Free PMC article. Review.
-
Profilin is involved in G1 to S phase progression and mitotic spindle orientation during Leishmania donovani cell division cycle.PLoS One. 2022 Mar 22;17(3):e0265692. doi: 10.1371/journal.pone.0265692. eCollection 2022. PLoS One. 2022. PMID: 35316283 Free PMC article.
-
The Trypanosoma brucei RNA-binding protein DRBD18 ensures correct mRNA trans splicing and polyadenylation patterns.RNA. 2022 Sep;28(9):1239-1262. doi: 10.1261/rna.079258.122. Epub 2022 Jul 6. RNA. 2022. PMID: 35793904 Free PMC article.
-
Comparative proteomics of the two T. brucei PABPs suggests that PABP2 controls bulk mRNA.PLoS Negl Trop Dis. 2018 Jul 24;12(7):e0006679. doi: 10.1371/journal.pntd.0006679. eCollection 2018 Jul. PLoS Negl Trop Dis. 2018. PMID: 30040867 Free PMC article.
References
-
- Campbell D.A., Thomas S., Sturm N.R. Transcription in kinetoplastid protozoa: why be normal? Microbes Infect. 2003;5:1231–1240. - PubMed
-
- Hershey J.W.B., Merrick W.C. Pathway and mechanism of initiation of protein synthesis. In: Sonenberg N., Hershey J.W.B., Mathews M.B., editors. Translational Control Of Gene Expression. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2000. pp. 33–88.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

