Cell fate-specific regulation of EGF receptor trafficking during Caenorhabditis elegans vulval development
- PMID: 16688213
- PMCID: PMC1478196
- DOI: 10.1038/sj.emboj.7601137
Cell fate-specific regulation of EGF receptor trafficking during Caenorhabditis elegans vulval development
Abstract
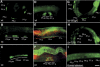
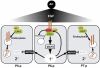
By controlling the subcellular localization of growth factor receptors, cells can modulate the activity of intracellular signal transduction pathways. During Caenorhabditis elegans vulval development, a ternary complex consisting of the LIN-7, LIN-2 and LIN-10 PDZ domain proteins localizes the epidermal growth factor receptor (EGFR) to the basolateral compartment of the vulval precursor cells (VPCs) to allow efficient receptor activation by the inductive EGF signal from the anchor cell. We have identified EGFR substrate protein-8 (EPS-8) as a novel component of the EGFR localization complex that links receptor trafficking to cell fate specification. EPS-8 expression is upregulated in the primary VPCs, where it creates a positive feedback loop in the EGFR/RAS/MAPK pathway. The membrane-associated guanylate kinase LIN-2 recruits EPS-8 into the receptor localization complex to retain the EGFR on the basolateral plasma membrane, and thus allow maximal receptor activation in the primary cell lineage. Low levels of EPS-8 in the neighboring secondary VPCs result in the rapid degradation of the EGFR, allowing these cells to adopt the secondary cell fate. Extracellular signals thus regulate EGFR trafficking in a cell type-specific manner to control pattern formation during organogenesis.
Figures




References
-
- Berset T, Hoier EF, Battu G, Canevascini S, Hajnal A (2001) Notch inhibition of RAS signaling through MAP kinase phosphatase LIP-1 during C. elegans vulval development. Science 291: 1055–1058 - PubMed
-
- Burdine RD, Branda CS, Stern MJ (1998) EGL-17(FGF) expression coordinates the attraction of the migrating sex myoblasts with vulval induction in C. elegans. Development 125: 1083–1093 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

