Cytotoxin ClyA from Escherichia coli assembles to a 13-meric pore independent of its redox-state
- PMID: 16688219
- PMCID: PMC1478193
- DOI: 10.1038/sj.emboj.7601130
Cytotoxin ClyA from Escherichia coli assembles to a 13-meric pore independent of its redox-state
Abstract
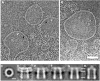
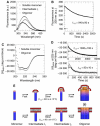
ClyA is a pore-forming toxin from virulent Escherichia coli and Salmonella enterica strains. Here, we show that the intrinsic hemolytic activity of ClyA is independent of its redox state, and that the assembly of both reduced and oxidized ClyA to the ring-shaped oligomer is triggered by contact with lipid or detergent. A rate-limiting conformational transition in membrane-bound ClyA monomers precedes their assembly to the functional pore. We obtained a three-dimensional model of the detergent-induced oligomeric complex at 12 A resolution by combining cryo- and negative stain electron microscopy with mass measurements by scanning transmission electron microscopy. The model reveals that 13 ClyA monomers assemble into a cylinder with a hydrophobic cap region, which may be critical for membrane insertion.
Figures





References
-
- Atkins A, Wyborn NR, Wallace AJ, Stillman TJ, Black LK, Fielding AB, Hisakado M, Artymiuk PJ, Green J (2000) Structure–function relationships of a novel bacterial toxin, hemolysin E. The role of alpha G. J Biol Chem 275: 41150–41155 - PubMed
-
- Braun M, Silhavy TJ (2002) Imp/OstA is required for cell envelope biogenesis in Escherichia coli. Mol Microbiol 45: 1289–1302 - PubMed
-
- del Castillo FJ, Leal SC, Moreno F, del Castillo I (1997) The Escherichia coli K-12 sheA gene encodes a 34-kDa secreted haemolysin. Mol Microbiol 25: 107–115 - PubMed
-
- del Castillo FJ, Moreno F, del Castillo I (2001) Secretion of the Escherichia coli K-12 SheA hemolysin is independent of its cytolytic activity. FEMS Microbiol Lett 204: 281–285 - PubMed
-
- Choe S, Bennett MJ, Fujii G, Curmi PM, Kantardjieff KA, Collier RJ, Eisenberg D (1992) The crystal structure of diphtheria toxin. Nature 357: 216–222 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

