The ESCRT complexes: structure and mechanism of a membrane-trafficking network
- PMID: 16689637
- PMCID: PMC1648078
- DOI: 10.1146/annurev.biophys.35.040405.102126
The ESCRT complexes: structure and mechanism of a membrane-trafficking network
Abstract
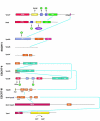
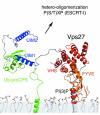
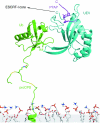
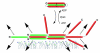
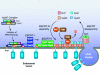
The ESCRT complexes and associated proteins comprise a major pathway for the lysosomal degradation of transmembrane proteins and are critical for receptor downregulation, budding of the HIV virus, and other normal and pathological cell processes. The ESCRT system is conserved from yeast to humans. The ESCRT complexes form a network that recruits monoubiquitinated proteins and drives their internalization into lumenal vesicles within a type of endosome known as a multivesicular body. The structures and interactions of many of the components have been determined over the past three years, revealing mechanisms for membrane and cargo recruitment and for complex assembly.
Figures


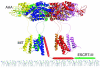
References
-
- Amerik AY, Li SJ, Hochstrasser M. Analysis of the deubiquitinating enzymes of the yeast Saccharomyces cerevisiae. Biol. Chem. 2000;381:981. - PubMed
-
- Antonyak MA, McNeill CJ, Wakshlag JJ, Boehm JE, Cerione RA. Activation of the Ras-ERK pathway inhibits retinoic acid-induced stimulation of tissue transglutaminase expression in NIH3T3 cells. J. Biol. Chem. 2003;278:15859. - PubMed
-
- Asao H, Sasaki Y, Arita T, Tanaka N, Endo K, et al. Hrs is associated with STAM, a signal-transducing adaptor molecule - Its suppressive effect on cytokine-induced cell growth. J. Biol. Chem. 1997;272:32785. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
