Novel genes encoding six kinds of three-finger toxins in Ophiophagus hannah (king cobra) and function characterization of two recombinant long-chain neurotoxins
- PMID: 16689684
- PMCID: PMC1550305
- DOI: 10.1042/BJ20060004
Novel genes encoding six kinds of three-finger toxins in Ophiophagus hannah (king cobra) and function characterization of two recombinant long-chain neurotoxins
Abstract
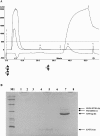
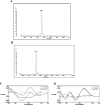
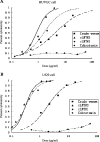
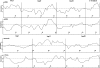
Three-finger toxins are a family of low-molecular-mass toxins (<10 kDa) having very similar three-dimensional structures. In the present study, 19 novel cDNAs coding three-finger toxins were cloned from the venom gland of Ophiophagus hannah (king cobra). Alignment analysis showed that the putative peptides could be divided into six kinds of three-finger toxins: LNTXs (long-chain neurotoxins), short-chain neurotoxins, cardiotoxins (CTXs), weak neurotoxins, muscarinic toxins and a toxin with a free SH group. Furthermore, a phylogenetic tree was established on the basis of the toxin cDNAs and the previously reported similar nucleotide sequences from the same source venom. It indicated that three-finger-toxin genes in O. hannah diverged early in the course of evolution by long- and short-type pathways. Two LNTXs, namely rLNTX1 (recombinant LNTX1) and rLNTX3, were expressed and showed cytolytic activity in addition to their neurotoxic function. By comparing the functional residues, we offer some possible explanations for the differences in their neurotoxic function. Moreover, a plausible elucidation of the additonal cytolytic activity was achieved by hydropathy-profile analysis. This, to our knowledge, is the first observation that recombinant long chain alpha-neurotoxins have a CTX-like cytolytic activity.
Figures




References
-
- Ricciardi A., le Du M.-H., Khayati M., Dajas F., Jean-Claude B., Menez A., Ducancel F. Do structural deviations between toxins adopting the same fold reflect functional differences? J. Biol. Chem. 2000;275:18302–18310. - PubMed
-
- Hall Z. W. α-Neurotoxins and their relatives: foes and friends? Neuron. 1999;23:4–5. - PubMed
-
- Jerusalinsky D., Harvey A. L. Toxins from mamba venoms: small proteins with selectivities for different subtypes of muscarinic acetylcholine receptors. Trends Pharmacol. Sci. 1994;15:424–430. - PubMed
-
- Utkin Y. N., Kukhtina V. V., Kryukva E. V., Chiodini F., Bertrand D., Methfessel C., Tsetlin V. I. “Weak toxin” from Naja kaouthia is a nontoxic antagonist of α7 and muscle-type nicotinic acetylcholine receptors. J. Biol. Chem. 2001;276:15810–15815. - PubMed
-
- Harvey A. L. Cardiotoxins from cobra venoms: possible mechanism of action. J. Toxicol. Toxin Rev. 1985;4:41–69.
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

