Fine-scale time-lapse analysis of the biphasic, dynamic behaviour of the two Vibrio cholerae chromosomes
- PMID: 16689793
- PMCID: PMC2779472
- DOI: 10.1111/j.1365-2958.2006.05175.x
Fine-scale time-lapse analysis of the biphasic, dynamic behaviour of the two Vibrio cholerae chromosomes
Abstract
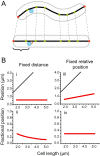
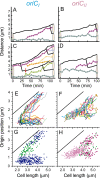
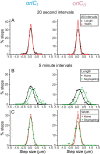
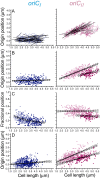
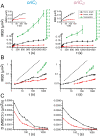
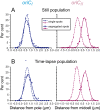
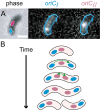
Using fluorescent repressor-operator systems in live cells, we investigated the dynamic behaviour of chromosomal origins in Vibrio cholerae, whose genome is divided between two chromosomes. We have developed a method of analysing fine-scale motion in the curved co-ordinate system of vibrioid bacteria. Using this method, we characterized two different modes of chromosome behaviour corresponding to periods between segregation events and periods of segregation. Between segregation events, the origin positions are not fixed but rather maintained within ellipsoidal caged domains, similar to eukaryotic interphase chromosome territories. These domains are approximately 0.4 microm wide and 0.6 microm long, reflecting greater restriction in the short axis of the cell. During segregation, movement is directionally biased, speed is comparable between origins, and cell growth can account for nearly 20% of the motion observed. Furthermore, the home domain of each origin is positioned by a different mechanism. Specifically, the oriC(I) domain is maintained at a constant actual distance from the pole regardless of cell length, while the oriC(II) domain is maintained at a constant relative position. Thus the actual position of oriC(II) varies with cell length. While the gross behaviours of the two origins are distinct, their fine-scale dynamics are remarkably similar, indicating that both experience similar microenvironments.
Figures

References
-
- Abuladze NK, Gingery M, Tsai J, Eiserling FA. Tail length determination in bacteriophage T4. Virology. 1994;199:301–310. - PubMed
-
- Berg HC. Random Walks in Biology. Princeton, NJ: Princeton University Press; 1993.
-
- Ebersbach G, Gerdes K. Bacterial mitosis: partitioning protein ParA oscillates in spiral-shaped structures and positions plasmids at mid-cell. Mol Microbiol. 2004;52:385–398. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

