Toward the structural genomics of complexes: crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis
- PMID: 16690741
- PMCID: PMC1472429
- DOI: 10.1073/pnas.0602606103
Toward the structural genomics of complexes: crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis
Abstract
The developing science called structural genomics has focused to date mainly on high-throughput expression of individual proteins, followed by their purification and structure determination. In contrast, the term structural biology is used to denote the determination of structures, often complexes of several macromolecules, that illuminate aspects of biological function. Here we bridge structural genomics to structural biology with a procedure for determining protein complexes of previously unknown function from any organism with a sequenced genome. From computational genomic analysis, we identify functionally linked proteins and verify their interaction in vitro by coexpression/copurification. We illustrate this procedure by the structural determination of a previously unknown complex between a PE and PPE protein from the Mycobacterium tuberculosis genome, members of protein families that constitute approximately 10% of the coding capacity of this genome. The predicted complex was readily expressed, purified, and crystallized, although we had previously failed in expressing individual PE and PPE proteins on their own. The reason for the failure is clear from the structure, which shows that the PE and PPE proteins mate along an extended apolar interface to form a four-alpha-helical bundle, where two of the alpha-helices are contributed by the PE protein and two by the PPE protein. Our entire procedure for the identification, characterization, and structural determination of protein complexes can be scaled to a genome-wide level.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures
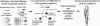



References
-
- Uetz P., Giot L., Cagney G., Mansfield T. A., Judson R. S., Knight J. R., Lockshon D., Narayan V., Srinivasan M., Pochart P., et al. Nature. 2000;403:623–627. - PubMed
-
- Gavin A. C., Bosche M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J. M., Michon A. M., Cruciat C. M., et al. Nature. 2002;415:141–147. - PubMed
-
- Ho Y., Gruhler A., Heilbut A., Bader G. D., Moore L., Adams S. L., Millar A., Taylor P., Bennett K., Boutilier K., et al. Nature. 2002;415:180–183. - PubMed
-
- Rual J. F., Venkatesan K., Hao T., Hirozane-Kishikawa T., Dricot A., Li N., Berriz G. F., Gibbons F. D., Dreze M., Ayivi-Guedehoussou N., et al. Nature. 2005;437:1173–1178. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

