Template-based coiled-coil antigens elicit neutralizing antibodies to the SARS-coronavirus
- PMID: 16697221
- PMCID: PMC7129695
- DOI: 10.1016/j.jsb.2006.03.019
Template-based coiled-coil antigens elicit neutralizing antibodies to the SARS-coronavirus
Abstract
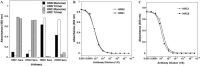
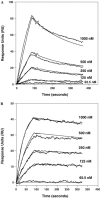
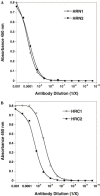
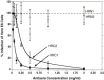
The Spike (S) glycoprotein of coronaviruses (CoV) mediates viral entry into host cells. It contains two hydrophobic heptad repeat (HR) regions, denoted HRN and HRC, which oligomerize the S glycoprotein into a trimer in the native state and when activated collapse into a six-helix bundle structure driving fusion of the host and viral membranes. Previous studies have shown that peptides of the HR regions can inhibit viral infectivity. These studies imply that the HR regions are accessible and that agents which can interact with them may prevent viral entry. In the present study, we have investigated an approach to generate antibodies that specifically recognize the HRN and HRC regions of the SARS-CoV spike (S) glycoprotein in order to evaluate whether these antibodies can inhibit viral infectivity and thus neutralize the SARS-CoV. In this regard, we incorporated HRN and HRC coiled-coil surface residues into a de novo designed two-stranded alpha-helical coiled-coil template for generating conformation-specific antibodies that recognize alpha-helices in proteins (Lu, S.M., Hodges, R.S., 2002. J. Biol. Chem. 277, 23515-23524). Eighteen surface residues from two regions of HRN and HRC were incorporated into the template and used to generate four anti-sera, HRN1, HRN2, HRC1, and HRC2. Our results show that all of the elicited anti-sera can specifically recognize HRN or HRC peptides and the native SARS-CoV S protein in an ELISA format. Flow cytometry (FACS) analysis, however, showed only HRC1 and HRC2 anti-sera could bind to native S protein expressed on the cell surface of Chinese hamster ovary cells, i.e., the cell surface structure of the S glycoprotein precluded the ability of the HRN1 or HRN2 anti-sera to see their respective epitope sites. In in vitro viral infectivity assays, no inhibition was observed for either HRN1 or HRN2 anti-serum, whereas both HRC1 and HRC2 anti-sera could inhibit SARS-CoV infection in a dose-dependent manner. Interestingly, the HRC1 anti-serum, which was a more effective inhibitor of viral infectivity compared to HRC2 anti-serum, could only bind the pre-fusogenic state of HRC, i.e., the HRC1 anti-serum did not recognize the six-helix bundle conformation (fusion state) whereas HRC2 anti-serum did. These results suggest that antibodies that are more specific for the pre-fusogenic state of HRC may be better neutralizing antibodies. Overall, these results clearly demonstrate that the two-stranded coiled-coil template acts as an excellent presentation system for eliciting helix-specific antibodies against highly conserved viral antigens and HRC1 and HRC2 peptides may represent potential candidates for use in a peptide vaccine against the SARS-CoV.
Figures




Similar articles
-
Biophysical characterization of HRC peptide analogs interaction with heptad repeat regions of the SARS-coronavirus Spike fusion protein core.J Struct Biol. 2006 Aug;155(2):162-75. doi: 10.1016/j.jsb.2006.03.024. Epub 2006 Apr 27. J Struct Biol. 2006. PMID: 16765058 Free PMC article.
-
Synthetic peptide studies on the severe acute respiratory syndrome (SARS) coronavirus spike glycoprotein: perspective for SARS vaccine development.Clin Chem. 2004 Jun;50(6):1036-42. doi: 10.1373/clinchem.2003.029801. Epub 2004 Mar 25. Clin Chem. 2004. PMID: 15044316 Free PMC article.
-
Receptor-binding domain of severe acute respiratory syndrome coronavirus spike protein contains multiple conformation-dependent epitopes that induce highly potent neutralizing antibodies.J Immunol. 2005 Apr 15;174(8):4908-15. doi: 10.4049/jimmunol.174.8.4908. J Immunol. 2005. PMID: 15814718
-
Vaccine design for severe acute respiratory syndrome coronavirus.Viral Immunol. 2005;18(2):327-32. doi: 10.1089/vim.2005.18.327. Viral Immunol. 2005. PMID: 16035944 Review.
-
The SARS-CoV S glycoprotein.Cell Mol Life Sci. 2004 Oct;61(19-20):2428-30. doi: 10.1007/s00018-004-4257-y. Cell Mol Life Sci. 2004. PMID: 15526150 Free PMC article. Review.
Cited by
-
Plasmodium vivax antigen discovery based on alpha-helical coiled coil protein motif.PLoS One. 2014 Jun 24;9(6):e100440. doi: 10.1371/journal.pone.0100440. eCollection 2014. PLoS One. 2014. PMID: 24959747 Free PMC article.
-
Role of plasmonics in detection of deadliest viruses: a review.Eur Phys J Plus. 2021;136(6):675. doi: 10.1140/epjp/s13360-021-01657-9. Epub 2021 Jun 20. Eur Phys J Plus. 2021. PMID: 34178567 Free PMC article. Review.
-
Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2.J Virol. 2010 Dec;84(24):12658-64. doi: 10.1128/JVI.01542-10. Epub 2010 Oct 6. J Virol. 2010. PMID: 20926566 Free PMC article.
-
Expression and characterization of recombinant S2 subunit of SARS-coronavirus S fusion protein.Adv Exp Med Biol. 2009;611:153-4. doi: 10.1007/978-0-387-73657-0_69. Adv Exp Med Biol. 2009. PMID: 19400136 Free PMC article. No abstract available.
-
Peptide nanoparticles as novel immunogens: design and analysis of a prototypic severe acute respiratory syndrome vaccine.Chem Biol Drug Des. 2009 Jan;73(1):53-61. doi: 10.1111/j.1747-0285.2008.00746.x. Chem Biol Drug Des. 2009. PMID: 19152635 Free PMC article.
References
-
- Bosch B.J., Martina B.E., Van Der Zee R., Lepault J., Haijema B.J., Versluis C., Heck A.J., De Groot R., Osterhaus A.D., Rottier P.J. Severe acute respiratory syndrome coronavirus (SARS-CoV) infection inhibition using spike protein heptad repeat-derived peptides. Proc. Natl. Acad. Sci. USA. 2004;101:8455–8460. - PMC - PubMed
-
- Bukreyev A., Lamirande E.W., Buchholz U.J., Vogel L.N., Elkins W.R., St Claire M., Murphy B.R., Subbarao K., Collins P.L. Mucosal immunisation of African green monkeys (Cercopithecus aethiops) with an attenuated parainfluenza virus expressing the SARS coronavirus spike protein for the prevention of SARS. Lancet. 2004;363:2122–2127. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

