Viewing dynamic assembly of molecular complexes by multi-wavelength single-molecule fluorescence
- PMID: 16698779
- PMCID: PMC1563747
- DOI: 10.1529/biophysj.106.084004
Viewing dynamic assembly of molecular complexes by multi-wavelength single-molecule fluorescence
Abstract
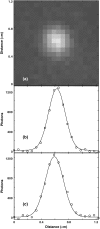
Complexes of macromolecules that transiently self-assemble, perform a particular function, and then dissociate are a recurring theme in biology. Such systems often have a large number of possible assembly/disassembly intermediates and complex, highly branched reaction pathways. Measuring the single-step kinetic parameters in these reactions would help to identify the functionally significant pathways. We have therefore constructed a novel single-molecule fluorescence microscope capable of efficiently detecting the colocalization of multiple components in a macromolecular complex when each component is labeled using a different color fluorescent dye. In this through-objective excitation, total internal reflection instrument, the dichroic mirror conventionally used to spectrally segregate the excitation and emission pathways was replaced with small broadband mirrors. This design spatially segregates the excitation and emission pathways and thereby permits efficient collection of the spectral range of emitted fluorescence when three or more dyes are used. In a test experiment with surface-immobilized single-stranded DNA molecules, we directly monitored the time course of a hybridization reaction with three different oligonucleotides, each labeled with a different color dye. The experiment reveals which of the possible reaction intermediates were traversed by each immobilized molecule, measures the hybridization rate constants for each oligonucleotide, and characterizes kinetic interdependences of the reaction steps.
Figures







References
-
- Funatsu, T., Y. Harada, M. Tokunaga, K. Saito, and T. Yanagida. 1995. Imaging of single fluorescent molecules and individual ATP turnovers by single myosin molecules in aqueous solution. Nature. 374:555–559. - PubMed
-
- Rasnik, I., S. Myong, W. Cheng, T. M. Lohman, and T. Ha. 2004. DNA-binding orientation and domain conformation of the E. coli rep helicase monomer bound to a partial duplex junction: single-molecule studies of fluorescently labeled enzymes. J. Mol. Biol. 336:395–408. - PubMed
-
- Blanchard, S. C., R. L. Gonzalez, H. D. Kim, S. Chu, and J. D. Puglisi. 2004. tRNA selection and kinetic proofreading in translation. Nat. Struct. Mol. Biol. 11:1008–1014. - PubMed
-
- Zhuang, X., L. E. Bartley, H. P. Babcock, R. Russell, T. Ha, D. Herschlag, and S. Chu. 2000. A single-molecule study of RNA catalysis and folding. Science. 288:2048–2051. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

