Minor groove deformability of DNA: a molecular dynamics free energy simulation study
- PMID: 16698780
- PMCID: PMC1563752
- DOI: 10.1529/biophysj.106.083816
Minor groove deformability of DNA: a molecular dynamics free energy simulation study
Abstract
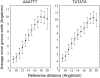
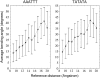
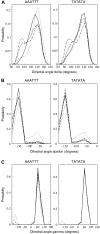
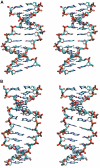
The conformational deformability of nucleic acids can influence their function and recognition by proteins. A class of DNA binding proteins including the TATA box binding protein binds to the DNA minor groove, resulting in an opening of the minor groove and DNA bending toward the major groove. Explicit solvent molecular dynamics simulations in combination with the umbrella sampling approach have been performed to investigate the molecular mechanism of DNA minor groove deformations and the indirect energetic contribution to protein binding. As a reaction coordinate, the distance between backbone segments on opposite strands was used. The resulting deformed structures showed close agreement with experimental DNA structures in complex with minor groove-binding proteins. The calculated free energy of minor groove deformation was approximately 4-6 kcal mol(-1) in the case of a central TATATA sequence. A smaller equilibrium minor groove width and more restricted minor groove mobility was found for the central AAATTT and also a significantly ( approximately 2 times) larger free energy change for opening the minor groove. The helical parameter analysis of trajectories indicates that an easier partial unstacking of a central TA versus AT basepair step is a likely reason for the larger groove flexibility of the central TATATA case.
Figures






References
-
- Hagerman, P. J. 1990. Flexibility of DNA. Annu. Rev. Biochem. 59:755–781. - PubMed
-
- El Hassan, M. A., and C. R. Calladine. 1995. The assessment of the geometry of dinucleotide steps in double-helical DNA: a new local calculation scheme. J. Mol. Biol. 251:648–664. - PubMed
-
- Packer, M. J., M. P. Dauncey, and C. A. Hunter. 2000. Sequence-dependent DNA structure: tetranucleotide conformational maps. J. Mol. Biol. 295:85–103. - PubMed
-
- Rhodes, D., J. W. Schwabe, L. Chapman, and L. Fairall. 1996. Towards an understanding of protein-DNA recognition. Philos. Trans. R. Soc. Lond. B Biol. Sci. 351:501–509. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

