Tryptophan rotamers as evidenced by X-ray, fluorescence lifetimes, and molecular dynamics modeling
- PMID: 16698786
- PMCID: PMC1563760
- DOI: 10.1529/biophysj.106.085100
Tryptophan rotamers as evidenced by X-ray, fluorescence lifetimes, and molecular dynamics modeling
Abstract
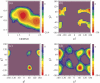
We investigated the native-state dynamics of the Bacillus caldolyticus cold-shock protein mutant Bc-Csp L66E, using fluorescence and appropriate molecular dynamics methods. Two fluorescence lifetimes were found, the amplitudes of which agree very well with tryptophan rotamer populations, obtained from parallel tempering calculations. Rotamer lifetimes were predicted by transition-state theory from high-temperature simulations. Transition pathways were extracted from the transition rates between individual rotameric states. The molecular dynamics also reveal the loop fluctuations in the native state.
Figures




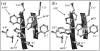
References
-
- Osterberg, F., G. M. Morris, M. F. Sanner, A. J. Olson, and D. S. Goodsell. 2002. Automated docking to multiple target structures: incorporation of protein mobility and structural water heterogeneity in AutoDock. Proteins. 46:34–40. - PubMed
-
- Stapley, B. J., and A. J. Doig. 1997. Free energies of amino acid side-chain rotamers in α-helices, β-sheets and α-helix N-caps. J. Mol. Biol. 272:456–464. - PubMed
-
- Lindorff-Larsen, K., P. Rogen, E. Paci, M. Vendruscolo, and C. M. Dobson. 2005. Protein folding and the organization of the protein topology universe. Trends Biochem. Sci. 30:13–19. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

