Strong physical constraints on sequence-specific target location by proteins on DNA molecules
- PMID: 16698961
- PMCID: PMC3303175
- DOI: 10.1093/nar/gkl271
Strong physical constraints on sequence-specific target location by proteins on DNA molecules
Abstract
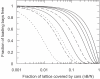
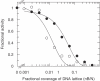
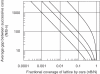
Sequence-specific binding to DNA in the presence of competing non-sequence-specific ligands is a problem faced by proteins in all organisms. It is akin to the problem of parking a truck at a loading bay by the side of a road in the presence of cars parked at random along the road. Cars even partially covering the loading bay prevent correct parking of the truck. Similarly on DNA, non-specific ligands interfere with the binding and function of sequence-specific proteins. We derive a formula for the probability that the loading bay is free from parked cars. The probability depends on the size of the loading bay and allows an estimation of the size of the footprint on the DNA of the sequence-specific protein by assaying protein binding or function in the presence of increasing concentrations of non-specific ligand. Assaying for function gives an 'activity footprint'; the minimum length of DNA required for function rather than the more commonly measured physical footprint. Assaying the complex type I restriction enzyme, EcoKI, gives an activity footprint of approximately 66 bp for ATP hydrolysis and 300 bp for the DNA cleavage function which is intimately linked with translocation of DNA by EcoKI. Furthermore, considering the coverage of chromosomal DNA by proteins in vivo, our theory shows that the search for a specific DNA sequence is very difficult; most sites are obscured by parked cars. This effectively rules out any significant role in target location for mechanisms invoking one-dimensional, linear diffusion along DNA.
Figures


Similar articles
-
Sequence-specific DNA binding by EcoKI, a type IA DNA restriction enzyme.J Mol Biol. 1998 Nov 13;283(5):963-76. doi: 10.1006/jmbi.1998.2143. J Mol Biol. 1998. PMID: 9799636
-
The DNA binding characteristics of the trimeric EcoKI methyltransferase and its partially assembled dimeric form determined by fluorescence polarisation and DNA footprinting.J Mol Biol. 1998 Nov 13;283(5):947-61. doi: 10.1006/jmbi.1998.2142. J Mol Biol. 1998. PMID: 9799635
-
Combinatorial determination of sequence specificity for nanomolar DNA-binding hairpin polyamides.Biochemistry. 2003 Jun 10;42(22):6891-903. doi: 10.1021/bi027373s. Biochemistry. 2003. PMID: 12779344
-
Footprinting: a method for determining the sequence selectivity, affinity and kinetics of DNA-binding ligands.Methods. 2007 Jun;42(2):128-40. doi: 10.1016/j.ymeth.2007.01.002. Methods. 2007. PMID: 17472895 Review.
-
How to get from A to B: strategies for analysing protein motion on DNA.Eur Biophys J. 2002 Jul;31(4):257-67. doi: 10.1007/s00249-002-0224-4. Epub 2002 May 30. Eur Biophys J. 2002. PMID: 12122472 Review.
Cited by
-
Structural Basis of Enhanced Facilitated Diffusion of DNA-Binding Protein in Crowded Cellular Milieu.Biophys J. 2020 Jan 21;118(2):505-517. doi: 10.1016/j.bpj.2019.11.3388. Epub 2019 Nov 29. Biophys J. 2020. PMID: 31862109 Free PMC article.
-
The effects of transcription factor competition on gene regulation.Front Genet. 2013 Oct 7;4:197. doi: 10.3389/fgene.2013.00197. eCollection 2013. Front Genet. 2013. PMID: 24109486 Free PMC article.
-
Sliding and jumping of single EcoRV restriction enzymes on non-cognate DNA.Nucleic Acids Res. 2008 Jul;36(12):4118-27. doi: 10.1093/nar/gkn376. Epub 2008 Jun 10. Nucleic Acids Res. 2008. PMID: 18544605 Free PMC article.
-
The influence of transcription factor competition on the relationship between occupancy and affinity.PLoS One. 2013 Sep 27;8(9):e73714. doi: 10.1371/journal.pone.0073714. eCollection 2013. PLoS One. 2013. PMID: 24086290 Free PMC article.
-
Optimal Length of Conformational Transition Region in Protein Search for Targets on DNA.J Phys Chem Lett. 2017 Sep 7;8(17):4049-4054. doi: 10.1021/acs.jpclett.7b01750. Epub 2017 Aug 15. J Phys Chem Lett. 2017. PMID: 28796515 Free PMC article.
References
-
- Pettijohn, D.E. 1982. Structure and properties of the bacterial nucleoid Cell 30667–669 - PubMed
-
- Neidhardt, F.C., Ingraham, J.L., Schaechter, M. Physiology of the Bacterial Cell: A Molecular Approach 1990. Sunderland MA Sinauer Assoc
-
- Pollard, T. and Earnshaw, W. Cell Biology 2002. Philadelphia Saunders

